#13,154
About a month ago, in Russia As An Outlier in This Year's Flu Epidemic, I wrote that even as North America, Europe, and Asia were all reporting heavy outbreaks of influenza, as of Epi Week 2 Russia had yet to cross the epidemic threshold, coming closest (under by 1.4%) in early December (week 51).
A bit out of the ordinary, as we saw reported six weeks ago in Eurosurveillance: Changes In Timing Of Influenza Epidemics - WHO European Region 1996-2016, over the past 2 decades Russia's flu seasons have tended to peak earlier with each passing year.
Every flu season is different, and nothing is set in stone.While flu in Russia receded from its initial peak in early December flu activity has begun to rise again over the past couple of weeks, and in the latest (week 6) flu surveillance report from the Russian Institute of Influenza, their rate of influenza has finally crossed the epidemic threshold.
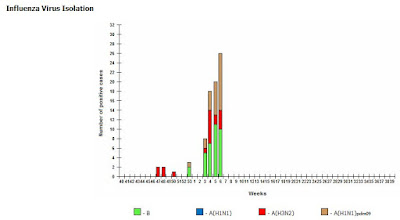
Since our last look at epi week two, H1N1pdm09 has surged (see chart below), while H3N2 has decreased. Influenza B continues strong, making up about 35% of samples tested.
Some excerpts from the Week 6 Russian Epidemic Situation Report, including reports of 3 H1N1 viruses in Moscow showing signs of resistance to NAI (Neuraminidase Inhibiting) antiviral drugs. I'll return with a bit more antiviral resistance.
Week 05.02.2018-11.02.2018
Influenza and ARI morbidity data
Epidemiological data show increase of influenza and other ARI activity in Russia in comparison with previous week. The ILI & ARI incidence rate (77.5 per 10 000 of population) was above by 6.8% the new nationalwide baseline (72.6) calculated by RII NIC for 2017-2018 season.
ILI and ARI epidemic thresholds were exceeded in 6 of 61 cities collaborating with two WHO NICs in Russia.
Conclusion
Influenza and ARI morbidity data. Increase of influenza and other ARI activity was registered during week 06.2018 in Russia. The ILI & ARI incidence rate (77.5 per 10 000 of population) was above by 6.8% the national wide baseline.
Etiology of ILI & ARI morbidity. The overall percent of respiratory samples positive for influenza was estimated as 10.0%. Proportion of influenza A(H1N1)pdm09, A(H3N2) and B viruses was estimated as 29.0%, 35.1% and 35.5%, respectively.
Antigenic characterization. 39 influenza viruses were characterized antigenically in two NICs, including 14 influenza A(H1N1)pdm09 viruses, 7 influenza A(H3N2) strains and 18 influenza type B strains. All influenza A(H1N1)pdm09 and A(H3N2) strains matched influenza vaccine strains for the season 2017-2018. 15 influenza type B strains of Yamagata lineage were like B/Phuket/3073/2013 reference virus, 3 influenza type B strains of Victoria lineage were antigenically related to B/Brisbain/60/2008 virus.
Genetic characterization. Full-genome NGS of 58 influenza positive samples and viruses from 6 cities was conducted. 16 influenza A(H1N1)pdm09 viruses belonged to phylogenetic group 6B.1 with amino acid substitutions in HA S84N, S162N and I216T.
According to phylogenetic analisis of HA 18 of 22 tested influenza A(H3N2) viruses belonged to clade 3C.2a carring aa substitutions L3I, N144S, F159Y, K160T, N225D and Q311H in HA1. Four influenza A(H3N2) viruses belonged to genetic subgroup 3C.2a1 and carried aa substitutions K92R, N121K, T135K and H311Q. 2 influenza B viruses of Victoria-lineage belonged to genetic subgroup 1A (B/Brisbane/60/2008-like). All 18 influenza B viruses of Yamagata-lineage belonged to clade 3 (B/Phuket/3073/2013-like) and had substitution L172Q and M251V in HA1.
Susceptibility to antivirals. Most viruses were susceptible to NA inhibitors excluding three influenza A(H1N1)pdm09 strains isolated in Moscow which had H275Y amino acid substitution in NA responsible for highly reduced susceptibility to oseltamivir and zanamivir.
14 influenza strains tested in MUNANA-assay for antiviral resistance to NA inhibitors in RII NIC, including 3 A(H1N1)pdm09 strains isolated in St.Petersburg, 4 A(H3N2), two B Victoria strains and 5 B Yamagata viruses were susceptible to oseltamivir and zanamivir. All influenza A strains tested were resistant to rimantadine.
(Continue . . . )Percent of positive ARI cases of non-influenza etiology (PIV, adeno- and RSV) was estimated as 24.7% of investigated patients by IFA and 16.7% by PCR. Last weeks RSV dominated among ARI agents.
In sentinel surveillance system clinical samples from 164 SARI and ILI/ARI patients were investigated by rRT-PCR. 10 (12.5%) influenza cases were detected among SARI patients, including 1 influenza A(H1N1)pdm09 case, 5 influenza A(H3N2) cases and 4 influenza B cases. Among ILI/ARI patients 28 (33.3%) influenza cases were detected, including 4 influenza A(H1N1)pdm09 cases, 14 influenza A(H3N2) cases and 10 influenza B cases.
During the 2008-2009 flu season (just before the 2009 H1N1 pandemic arrived) public health officials were scrambling because the old H1N1 virus had - in the space of a year - gone from showing about 1% resistance to oseltamivir (aka `Tamiflu') to being nearly 100% resistant.
The CDC was forced to issue major new guidance for the use of antivirals (see CIDRAP article With H1N1 resistance, CDC changes advice on flu drugs).This resistance was due to the acquisition of an H275Y mutation - where a single amino acid substitution (histidine (H) to tyrosine (Y)) occured at the neuraminidase position 275 (Note: some scientists use 'N2 numbering' (H274Y)).
Perhaps the one saving grace of the 2009 pandemic is that it supplanted the old H1N1 virus with a new one that was still susceptible to oseltamivir. We've been watching ever since then for any signs that the new pH1N1 virus has been gaining resistance, but for the most part, the news has been pretty good.
Rates of resistant pH1N1 viruses have remained low - around 1% - and like we saw prior to 2007, have generally been seen in (often immunocompromised) patients after they were placed on antivirals - due to `spontaneous mutations’.The most recent FluView report from the CDC reports testing 431 H1N1 viruses since Oct 1st 2017, and finding only 4 (0.9%) showing signs of resistance.
That said, we have seen a few worrisome instances of H1N1pdm viruses showing resistant to NAI antiviral drugs around the globe, including:
- In December of 2011, in NEJM: Oseltamivir Resistant H1N1 in Australia, we looked at a cluster of oseltamivir (Tamiflu ®) resistant H1N1 viruses in and around the Newcastle area of New South Wales.
- In 2014's Eurosurveillance: Community Cluster Of Antiviral Resistant pH1N1 in Japan, we looked at a cluster of six genetically similar resistant viruses in Sapporo, Japan - but without epidemiological links.
- Also in 2014, in PLoS Pathogens: Fitness Advantage From Permissive NA Mutations In Oseltamivir Resistant pH1N1, we saw a study that explored the potential of pH1N1 eventually following the same course as its predecessor.
- And in 2016, in Eurosurveilance: A(H1N1)pdm09 Virus With Cross-Resistance To Oseltamivir & Peramivir - Japan, March 2016 we looked at an elevated number of NAI resistant viruses with `permissive mutations' circulating in Japan.
That said, we don't have enough information to know whether these three resistant viruses reported from Moscow are significant or not.We don't know if any of these these cases are epidemiologically linked, if they occurred after treatment had begun (aka `spontaneous mutations'), how many viruses have been characterized this year in Moscow, or any other particulars.
Hopefully we'll get some additional information in next week's report.