# 5058
When it comes to avian influenzas, H5N1 gets the lion’s share of our attention, but it isn’t the only avian flu strain with the potential to jump to humans.
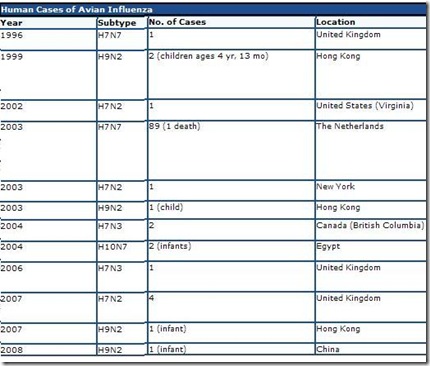
Below you’ll find a chart lifted and edited from CIDRAP’s excellent overview Avian Influenza (Bird Flu): Implications for Human Disease showing non-H5N1 avian flu infections in humans over the past decade.
Since surveillance is – at best - haphazard (or even non-existent) in many parts of the world, how often this really happens is unknown.
Like the novel swine flu cases that caused such a stir last week, they probably happen a bit more often than we realize.
A couple of years ago, we saw a study in PNAS that indicated that the H7 virus might be moving more towards adapting to humans.
You can read more about this in a couple of blogs from 2008, H7's Coming Out Party and H7 Study Available Online At PNAS.
Today, in a similar vein, we have study appearing in the Virology Journal that suggests that the H9N2 virus may also be evolving more towards humans as well.
While most virological research studies are in vivo or in vitro, this one is in silica, or based primarily on computer analysis of existing data.
Azeem M Butt, Samerene Siddique, Muhammad Idrees and Yigang Tong
Virology Journal 2010, 7:319 doi:10.1186/1743-422X-7-319
Published: 15 November 2010
Abstract (provisional)
Background
H9N2 avian influenza A viruses have become panzootic in Eurasia over the last decade and have caused several human infections in Asia since 1998. To study their evolution and zoonotic potential, we conducted an in silico analysis of H9N2 viruses that have infected humans between 1997 and 2009 and identified potential novel reassortments.
Results
A total of 22 hemagglutinin (HA) and neuraminidase (NA) nucleotide and deduced amino acid sequences were retrieved from the NCBI flu database.
It was identified that mature peptide sequences of HA genes isolated from humans in 2009 had glutamine at position 226 (H3) of the receptor binding site, indicating a preference to bind to the human alpha (2-6) sialic acid receptors, which is different from previously isolated viruses and studies where the presence of leucine at the same position contributes to preference for human receptors and presence of glutamine towards avian receptors.
Similarly, strains isolated in 2009 possessed new motif R-S-N-R in spite of typical R-S-S-R at the cleavage site of HA, which isn't reported before for H9N2 cases in humans. Other changes involved loss, addition, and variations in potential glycosylation sites as well as in predicted epitopes. The results of phylogenetic analysis indicated that HA and NA gene segments of H9N2 including those from current and proposed vaccine strains belong to two different Eurasian phylogenetic lineages confirming possible genetic reassortments.
Conclusions
These findings support the continuous evolution of avian H9N2 viruses towards human as host and are in favor of effective surveillance and better characterization studies to address this issue.
The complete article is available as a provisional PDF. The fully formatted PDF and HTML versions are in production.
In December 2008, after receiving the news of a baby in Hong Kong having been diagnosed with H9N2, I reran a blog featuring an interview in which world famous Hong Kong virologist Malik Peiris cautioned that the H9N2 virus may be circulating far more commonly than we believe.
Revisiting A Malik Peiris Interview On H9N2
As it exists now, H9 poses a low threat to humans.
Sporadic reports of human infections – particularly when there is no evidence of ongoing transmission – are interesting, but not particularly alarming.
But H9, like a number of other avian viruses (H5’s, H7’s) have some pandemic potential, particularly if they can `drift’ or mutate sufficiently, or pick up genetic material from other viruses.
This `reassortment’ could conceivably create a new, hybrid strain of influenza.
How likely is this to happen?
Well, that’s the big question. No one really knows.
We just know that it is possible.
It obviously doesn’t happen often, otherwise we’d be hip deep in new, hybrid viruses all of the time. But this is essentially the route that the 2009 H1N1 virus took to become a pandemic, and is likely the way the 1957 and 1968 pandemics came about.
And so we watch these rare human cases with great interest. A dangerous reassortment or mutation may never occur with the H9 virus, or it could happen tomorrow.
Influenza viruses, as they say, are unpredictable.