#17,411
While influenza A viruses are believed to have originated in avian species (primarily waterfowl), over time they’ve managed to infect, and adapt to, a wide variety of avian and non-avian hosts.
Humans, obviously, and swine . . . but we’ve also seen influenza A viruses infect mink, bats, marine mammals, horses, small peridomestic mammals, and companion animals like cats and dogs. Many of these susceptibilities have only been confirmed in the past 20 years or so.
For the most part, these flu viruses have adapted to, and circulate within, a limited host-range. Only rarely do we see a swine-origin virus infect humans, or the spillover of an avian virus into a mammalian host.
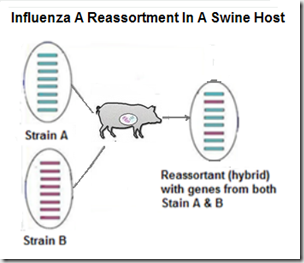
But influenza's superpower is its ability to evolve, and adapt to new hosts. Under the right conditions, these viruses are able to reassort (see graphic below), and produce new, and potentially more dangerous, subtypes or genotypes.Increased Public Health Threat of Avian-origin H3N2 Influenza Virus During Evolution in Dogs (Revisited)
One Health Adv.: Mink Infection With Influenza A Viruses - An Ignored Intermediate Host?
PrePrint: HPAI H5N1 Infections in Wild Red Foxes Show Neurotropism and Adaptive Virus Mutations
EID Journal: Zoonotic Threat of G4 Genotype Eurasian Avian-Like Swine Influenza A(H1N1) Viruses, China, 2020
Pathogens: Emergence and Characterization of a Novel Reassortant Canine Influenza Virus Isolated from Cats
Zoonotic Animal Influenza Virus and Potential Mixing Vessel Hosts
by
Elsayed M. Abdelwhab 1,* and Thomas C. Mettenleiter 2,*
Viruses 2023, 15(4), 980; https://doi.org/10.3390/v15040980 (registering DOI)
Received: 7 March 2023 / Revised: 5 April 2023 / Accepted: 14 April 2023 / Published: 16 April 2023
Abstract
Influenza viruses belong to the family Orthomyxoviridae with a negative-sense, single-stranded segmented RNA genome. They infect a wide range of animals, including humans. From 1918 to 2009, there were four influenza pandemics, which caused millions of casualties.Frequent spillover of animal influenza viruses to humans with or without intermediate hosts poses a serious zoonotic and pandemic threat. The current SARS-CoV-2 pandemic overshadowed the high risk raised by animal influenza viruses, but highlighted the role of wildlife as a reservoir for pandemic viruses.In this review, we summarize the occurrence of animal influenza virus in humans and describe potential mixing vessel or intermediate hosts for zoonotic influenza viruses. While several animal influenza viruses possess a high zoonotic risk (e.g., avian and swine influenza viruses), others are of low to negligible zoonotic potential (e.g., equine, canine, bat and bovine influenza viruses). Transmission can occur directly from animals, particularly poultry and swine, to humans or through reassortant viruses in “mixing vessel” hosts.To date, there are less than 3000 confirmed human infections with avian-origin viruses and less than 7000 subclinical infections documented. Likewise, only a few hundreds of confirmed human cases caused by swine influenza viruses have been reported. Pigs are the historic mixing vessel host for the generation of zoonotic influenza viruses due to the expression of both avian-type and human-type receptors. Nevertheless, there are a number of hosts which carry both types of receptors and can act as a potential mixing vessel host. High vigilance is warranted to prevent the next pandemic caused by animal influenza viruses.
Our track record of predicting which zoonotic virus will spark the next pandemic is abysmal, which is why we follow so many threats. And still - as we saw with COVID-19 and the 2009 H1N1 pandemic - we often get hit by something unexpected from out of left field.(SNIP)
Potential “Mixing Vessel” Hosts
Mixing vessel hosts are those in which co-infection of two (or more) IAVs can occur simultaneously, leading to the potential for reassortment and generation of new IAV genotypes/phenotypes. They act as intermediate hosts for the spread of IAV between/to mammals, including humans. Although several host factors are incriminated in the ability of animal influenza to replicate in human cells, virus receptors on the cells are a major determinant of host susceptibility to influenza viruses and thus play an important role in infection and virulence.Sialic acid (SA) α-linked at C2 to galactose of a cellular glycoprotein or glycolipid is the most common receptor for influenza viruses. hIAV typically prefers binding to an α2,6-linked SA (galactose C6, designated hereafter as α2-6-SA) and avian IAV to an α2,3-linked (galactose C3, α2,3-SA) [206]. α2,6-SA in the respiratory tract is commonly referred to as the “human receptor” and α2,3-SA is found in the intestinal tract of birds and is referred to as the “avian receptor”. It was originally thought that humans exclusively express α2,6-SA, birds only express α2,3-SA, and pigs have both avian and human receptor types. Therefore, they play a role as a mixing vessel for the generation of different avian and human reassortants.New studies have changed this paradigm of species and tissue distribution of SA. Many mammalian and avian species possess both types of SA receptors with variable abundance, are susceptible to hIAV and AIV infection, and can play a role as mixing vessels, similar to pigs (Figure 3). As different methods have been used to identify SA receptors in understudied species, a direct comparison is not possible. Here, we summarized potential “mixing vessel” based on the distribution of avian and human SA receptors, the number of animal-to-human IAV transmission events, the number of IAV subtypes, the number of animal populations, and the direct and long contact with humans, and the severity of the disease (not dead end hosts) to “high probability”, including humans, pigs, minks, ferrets, seals, dogs, cats, and birds, particularly turkeys, chickens, quails, and ducks; “medium probability” mixing vessel hosts are non-human primates, raccoons, camels, pikas, horses, and zoo animals, including tigers and lions. The “low probability” hosts are foxes, bats, and whales (Figure 3).
Figure 3. Potential “mixing vessel” hosts for the generation of zoonotic animal influenza viruses. Potential mixing vessel hosts according to the frequency of infection, close contact with humans, the high number of populations, and the distribution of avian- and human-type receptors. Humans, pigs, minks, ferrets, seals, dogs, cats, and birds, particularly turkeys, chickens, quails, and ducks, are the “high probability” mixing vessel hosts; “medium probability” mixing vessel hosts are non-human primates, raccoons, camels, pikas, zoo animals, including tigers and lions, and horses. The “low probability” hosts for the generation of zoonotic animal IAV are foxes, bats, and whales.
(SNIP)
Summary and Concluding Remarks
Zoonotic pathogens are responsible for more than 60% of human infectious diseases [407]. Although several zoonotic viruses caused severe human casualties, including the current SARS-CoV2; influenza viruses were responsible for at least four confirmed pandemics in less than a century [408]. IAVs infect a wide range of host species. Avian and swine influenza viruses are of high zoonotic potential, while influenza viruses of bovine, equine, canine, and bat origin are of low zoonotic risk. Animal influenza viruses can transmit directly to humans without intermediate mammal hosts.
Beyond pigs, there are several potential mixing vessel hosts for the generation of zoonotic animal influenza viruses, including humans, minks, seals, dogs, cats, zoo animals, camels, and several species of birds. Given the extensive number of wild birds, poultry, swine, and companion animals (dogs and cats) and their close contact to an ever-increasing human population currently standing at eight billion, animal influenza viruses will remain a serious threat for public health. Migratory birds are the highly mobile reservoir for AIVs. Unlike the control of rabies in foxes, there is currently no technology to vaccinate or control IAV infection in the wild-bird reservoir. However, it is possible to limit the infections in domestic reservoirs through improved biosecurity measures, cost-effective culling strategies, and development and use of effective vaccines.
Measures are needed to protect non-human mammals (e.g., pigs, minks) from infection with hIAV and AIV and prevent the spread of AIV from and to wild birds. The recent incursion of zoonotic HPAIV H5Nx in wild birds is a game-changer [409]. The virus was transmitted over a long distance by migratory birds from Eurasia via Iceland to the American continent, reaching for the first time, South America [287]. This virus is highly virulent for domestic birds and is able to infect a wide-range of mammals, including humans. Thus, enhanced vigilance is required to monitor the spread and biological alterations of this virus which could develop into a new pandemic pathogen.
(Continue . . . )
While we may not be able to predict from what what direction the next pandemic will come, we can say with confidence that another pandemic will occur. They've been happening regularly for hundreds of years (see Pandemic Influenza's 500th Anniversary), and COVID-19 disproves the notion that `modern medicine' has somehow rendered them obsolete.
Eighteen months ago, in PNAS Research: Intensity and Frequency of Extreme Novel Epidemics, researchers suggested that the probability of novel disease outbreaks will likely grow three-fold in the next few decades.
Which is why we should be preparing seriously now, lest we risk being caught flat footed and unprepared.