#18,057
While we don't have any evidence of an ongoing spillover of H5N1 to humans in the United States, we have preprint (published yesterday) which documents the recent detection of H5N1 in wastewater collected from 9 (unnamed) Texas cities.
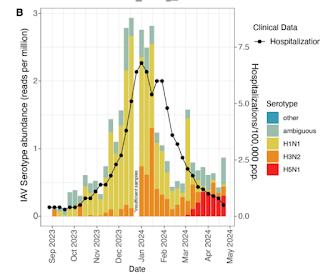
The first detections of H5N1 appeared in March (see chart above) and by April their rate exceeded that of seasonal influenza viruses.
The origins of these viruses are still thought to be primarily from avian or bovine sources, but some contributions from humans cannot be excluded.
I've posted the abstract and some brief excerpts below. Follow the link to read it in its entirety.
Virome Sequencing Identifies H5N1 Avian Influenza in Wastewater from Nine Cities
Michael J Tisza, Blake Hanson, Justin Ryan Clark, Li Wang, Katelyn Payne, Matthew C Ross, Kristina D Mena, Anna Gitter, Sara Joan Javornik Cregeen, Juwan J Cormier, Vasanthi J Avadhanula, Austen L Terwilliger, John E Balliew, Fuqing Wu, Janelle Rios, Jennifer Deegan, Pedro Piedra, Joseph F Petrosino, Eric Boerwinkle, Anthony W. Maresso
doi: https://doi.org/10.1101/2024.05.10.24307179
Preview PDF
Abstract
Avian influenza (serotype H5N1) is a highly pathogenic virus that emerged in domestic waterfowl in 1996. Over the past decade, zoonotic transmission to mammals, including humans, has been reported. Although human to human transmission is rare, infection has been fatal in nearly half of patients who have contracted the virus in past outbreaks. The increasing presence of the virus in domesticated animals raises substantial concerns that viral adaptation to immunologically naive humans may result in the next flu pandemic.
Wastewater-based epidemiology (WBE) to track viruses was historically used to track polio and has recently been implemented for SARS-CoV2 monitoring during the COVID-19 pandemic. Here, using an agnostic, hybrid-capture sequencing approach, we report the detection of H5N1 in wastewater in nine Texas cities, with a total catchment area population in the millions, over a two-month period from March 4th to April 25th, 2024.
Sequencing reads uniquely aligning to H5N1 covered all eight genome segments, with best alignments to clade 2.3.4.4b. Notably, 19 of 23 monitored sites had at least one detection event, and the H5N1 serotype became dominant over seasonal influenza over time. A variant analysis suggests avian or bovine origin but other potential sources, especially humans, could not be excluded. We report the value of wastewater sequencing to track avian influenza.
(SNIP)
In conclusion, we report the widespread detection of Influenza A H5N1 virus in wastewater from nine U.S. cities during the spring of 2024. Although the exact cause of the signal is currently unknown, lack of clinical burden along with genomic information suggests avian or bovine origin.
Given the now widespread presence of the virus in dairy cows, the concerning findings that unpasteurized milk may contain live virus, and that these two recent factors will increase the number of viral interactions with our species, wastewater monitoring should be readily considered as a sentinel surveillance tool that augments and accelerates our detection of evolutionary adaptations of significant concern.