#18,350
As the CDC graphic above illustrates, this summer's uptick in COVID activity - due to the emergence of the KP.3.1.1 variant last May - has begun to wane. The pattern with COVID, however, is that variants very rarely hold on to dominance more than 5 or 6 months, and the latest CDC Nowcast shows a new XEC variant is already making move to usurp the throne.
While KP.3.1.1 still holds on to a 57% share, XEC has been doubling every 2 weeks, growing from an estimated 2.3% a month ago in the United States, to 10.7% last week. At that rate, XEC could become dominant here by November.
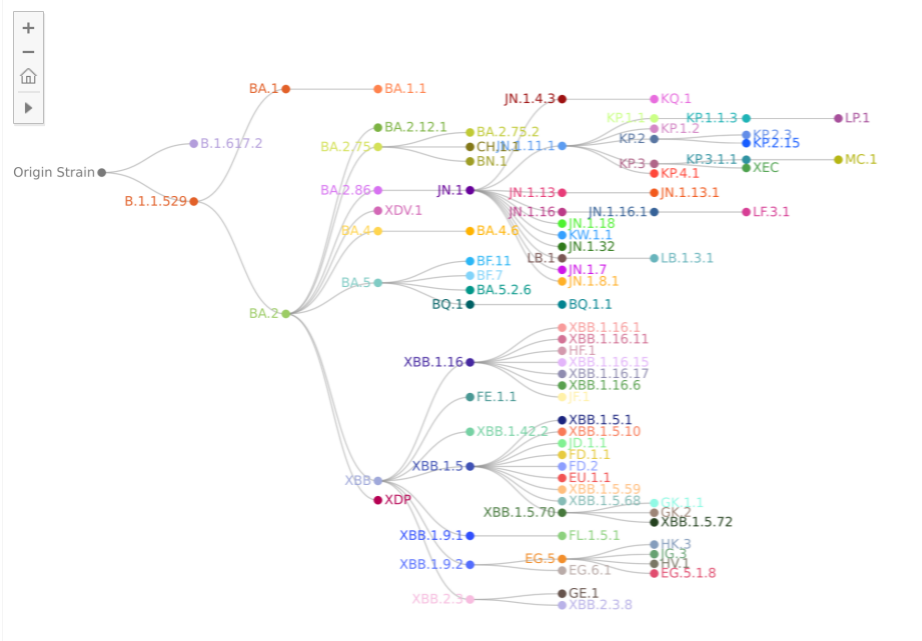
The SARS-CoV-2 virus continues to evolve and diversify (see diagram below), introducing new, often more `biologically fit', variants every few months.
On Thursday (Oct 17th) The Sato Lab (Kei Sato) @SystemsVirology published a preprint on the bioRxiv server characterizing this XEC variant, finding that it has the `right stuff' to make a run for global dominance in the next few months.
How long it will hold that position remains to be seen. I've reproduced the abstract below, so follow the link to read the full analysis.
Yu Kaku, Kaho Okumura, Shusuke Kawakubo, Keiya Uriu, Luo Chen, Yusuke Kosugi, Yoshifumi Uwamino, MST Monira Begum, Sharee Leong, Terumasa Ikeda, Kenji Sadamasu, Hiroyuki Asakura, Mami Nagashima, Kazuhisa Yoshimura, The Genotype to Phenotype Japan (G2P-Japan) Consortium, Jumpei Ito, Kei Satodoi: https://doi.org/10.1101/2024.10.16.618773
Abstract
The SARS-CoV-2 JN.1 variant (BA.2.86.1.1), arising from BA.2.86.1 with spike protein (S) substitution S:L455S, outcompeted the previously predominant XBB lineages by the beginning of 2024. Subsequently, JN.1 subvariants including KP.2 (JN.1.11.1.2) and KP.3 (JN.1.11.1.3), which acquired additional S substitutions (e.g., S:R346T, S:F456L, and S:Q493E), have emerged concurrently.
As of October 2024, KP.3.1.1 (JN.1.11.1.3.1.1), which acquired S:31del, outcompeted other JN.1 subvariants including KP.2 and KP.3 and is the most predominant SARS-CoV-2 variant in the world. Thereafter, XEC, a recombinant lineage of KS.1.1 (JN.13.1.1.1) and KP.3.3 (JN.1.11.1.3.3), was first identified in Germany on August 7, 2024. XEC acquired two S substitutions, S:T22N and S:F59S, compared with KP.3 through recombination, with a breakpoint at genomic position 21,738-22,599.
We estimated the relative effective reproduction number (Re) of XEC using a Bayesian multinomial logistic model based on genome surveillance data from the USA, the United Kingdom, France, Canada, and Germany, where this variant has spread as of August 2024.
In the USA, the Re of XEC is 1.13-fold higher than that of KP.3.1.1. Additionally, the other countries under investigation herein showed higher Re for XEC. These results suggest that XEC has the potential to outcompete the other major lineage including KP.3.1.1.
We then assessed the virological properties of XEC using pseudoviruses. Pseudovirus infection assay showed that the infectivity of KP.3.1.1 and XEC was significantly higher than that of KP.3. Although S:T22N did not affect the infectivity of the pseudovirus based on KP.3, S:F59S significantly increased it. Neutralization assay was performed using three types of human sera: convalescent sera after breakthrough infection (BTI) with XBB.1.5 or KP.3.3, and convalescent sera after JN.1 infection.
In all serum groups, XEC as well as KP.3.1.1 showed immune resistance when compared to KP.3 with statistically significant differences. In the cases of XBB.1.5 BTI sera and JN.1 infection sera, the 50% neutralization titers (NT50s) of XEC and KP.3.1.1 were comparable. However, we revealed that the NT50 of XEC was significantly (1.3-fold) lower than that of KP.3.1.1. Moreover, both S:T22N and S:F59S significantly (1.5-fold and 1.6-fold) increased the resistance to KP.3.3 BTI sera.
Here we showed that XEC exhibited higher pseudovirus infectivity and higher immune evasion than KP.3. Particularly, XEC exhibited more robust immune resistance to KP.3.3 BTI sera than KP.3.1.1. Our data suggest that the higher Re of XEC than KP.3.1.1 is attributed to this property and XEC will be a predominant SARS-CoV-2 variant in the world in the near future.
Despite reassurances (4 years ago) that we were only months away from achieving `herd immunity', and predictions the SARS-CoV-2 virus would eventually stabilize, and become a minor `seasonal' threat, COVID shows few signs of taking early retirement.
While XEC appears to be the latest contender for the viral throne, there are no guarantees another - more biologically `fit' variant - isn't already spreading somewhere in the world.