#18,346
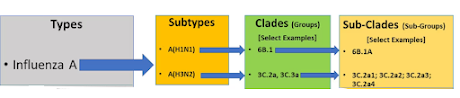
With 18 HA (hemagglutinin) types and 11 different NA (neuraminidase) types there are nearly 200 possible influenza subtypes (e.g. H1N1, H5N1, H7N8, etc.). To date, more than 130 have already been detected in nature.
Each subtype (based on the combination of HA and NA genes) is further classified by its HA clade (e.g. 2.3.4.4), and within each clade there can be multiple subclades (e.g. 2.3.4.4b, 2.3.2.1.c , etc.). This alone can yield thousands of variations.In North America alone, more than 100 genotypes have been identified over the past 3 years, with new ones destined to emerge over time. Globally, that number is much larger, although given the limits of surveillance and reporting, we aren't aware of all of them.
While the nomenclature for subtypes and clades have been standardized around the globe - different regions of the world have developed their own genotype classification system - making it difficult to compare studies or share data from Asia, and Europe, and North America.
Yesterday OFFLU - the WOAH/FAO joint network of expertise on avian influenza - released a statement proposing the creation of a cross-reference document this year, to be followed by `. . . a position paper that highlights the importance of a harmonized framework for addressing gaps in tracking, communication, and response to the evolving challenges posed by H5 viruses.'.
First the OFFLU statement, after which I'll have a brief postscript.
OFFLU Statement on the Development of a Global Consensus H5 Influenza Genotyping Framework
Since its inception in 2005, OFFLU (the WOAH-FAO network of expertise on animal influenza) has been closely monitoring the global impacts of avian influenza, including working with multiple countries and stakeholders affected by the current H5N1 HPAI panzootic. Field veterinarians and OFFLU scientists in FAO and WOAH designated influenza Reference Centres play a key role in responding to novel outbreaks and characterizing avian influenza (AI) viruses.
OFFLU is committed to ensuring effective stakeholder communication around animal influenzas. The World Organization of Health (WHO)/OFFLU H5 Evolution Working Group proposed a unified nomenclature1 specifically for high pathogenicity avian influenza (HPAI) H5 viruses of the goose/Guangdong lineage based upon the hemagglutinin gene only, which remains under continued revision. This nomenclature has successfully enabled the use of a universal, commonly understood system around the world by multiple stakeholders.
However, while viral genome sequences are increasingly available from diverse regions and hosts, the absence of a globally harmonized nomenclature for influenza A(H5) genotypes - particularly for highly pathogenic H5 goose/Guangdong viruses - poses a significant challenge to communication between animal and public health partners within and beyond the OFFLU network. In this context, genotypes are defined as viruses with a unique eight-gene segment composition resulting from reassortment events.
The current use of multiple genotyping systems impedes the tracking of genetic changes, hinders the identification of emerging strains, and complicates the assessment of virus spread and potential risks to animal and public health. To address this critical gap, OFFLU, in collaboration with global partners, is spearheading the development of a universal H5 influenza genotyping framework, beginning with the H5 goose/Guangdong lineage viruses, starting with clade 2.3.4.4b.
The Need for a Global Harmonized Genotyping Framework
Several tools and systems have been developed to classify and genotype animal influenza viruses, playing a crucial role in regional and local communication about avian influenza spread. While these approaches have significantly advanced the tracking and analysis of virus evolution, they remain continent or region-specific and lack global alignment. Given the unprecedented spread of avian influenza across geographic regions, there is a pressing need for a unified, widely accessible framework that builds on the strengths of existing systems to facilitate global communication. Importantly, this universal system will not replace regional systems but will serve as a means of global harmonization, ensuring interoperability without disrupting established practices. Such a framework would facilitate cross-border risk comparisons, enable efficient genotype tracing, and allow better communication regarding assessment of H5 wholegenome constellations, including the public health risk. Although creating and maintaining this framework presents significant challenges, it is a crucial step toward effectively confronting the evolving threats posed by avian influenza viruses.
OFFLU Approach
To develop the proposed genotyping framework, OFFLU Avian Influenza Technical Activity will first create a reference document, identifying and cross-referencing genotypes amongst existing classification systems from all continents, providing a foundation for standardization. This will be followed by a position paper that highlights the importance of a harmonized framework for addressing gaps in tracking, communication, and response to the evolving challenges posed by H5 viruses.
A technical team will develop a comprehensive genotyping framework to define all H5 goose/Guangdong lineage genotypes circulating globally, starting with HA clade 2.3.4.4b, and provide a nominal tool for cross-referencing genotypes across existing classification schemes from all continents. This framework will include a system for effectively communicating the significance of each genotype to decision-makers, ensuring clarity and avoiding misattribution or implications of country-specific origins. To promote inclusivity and relevance throughout this process, global experts and stakeholders in existing genotyping systems will be engaged and invited to contribute.
We plan to conduct initial testing of the framework and associated tools with global experts in late June 2025. We will then expand dissemination of the framework to scientists, policymakers, public health officials, and other stakeholders, emphasizing that the new genotype nomenclature is not an indicator of emerging variants but rather a universal naming system for improved communication. The framework will be publicly accessible, ensuring transparency and ease of use across regions and disciplines.
OFFLU (www.offlu.org) will continue to support the activities of its parent organisations (FAO and WOAH) and partners (WHO) in ensuring that scientifically sound information is available on strains of virus that are detected in animals.
Disclaimer: This statement provides the point of view of independent OFFLU experts and does not necessarily reflect the position of the parent organisations FAO and WOAH.
References
1 World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses. 2014 May;8(3):384-8. doi: 10.1111/irv.12230. Epub 2014 Jan 31. PMID: 24483237; PMCID: PMC4181488.
While there have been multiple genotypes of H5Nx going back decades - over the past 3 years, with the rapid global expansion of the virus - we've seen an unprecedented explosion in its diversity. Since different genotypes can possess different traits (transmissibility, host range, virulence, etc.), keeping track of them has become crucial.
The recently emerged H5N1 D1.1 genotype, for example, appears to produce more serious illness in humans than the `bovine' B3.13 genotype, while H5N6 viruses with recombinant internal genes derived from H9N2 have shown increased pathogenicity and transmissibility
Since this framework is to be geared to the HPAI H5 goose/Guangdong lineage of viruses (starting with clade clade 2.3.4.4b), it will also be useful for classifying other H5 subtypes, like H5N6, H5N5, and H5N8.
This is a big job, however, and it won't happen overnight. But is seems unlikely that HPAI H5 will be going away anytime soon.