# 5731
Salmonella infections – like most infectious diseases – go underreported in the United States and around the world. For many, infection is mild and often brief, and so medical treatment is never sought.
Those cases that do end up reported to the CDC usually represent only the tip of the iceberg – or in this case, a pyramid – of the actual number of cases.
Photo Credit CDC
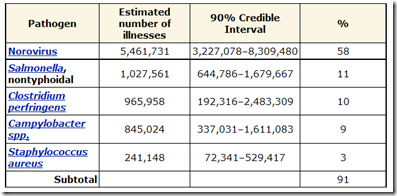
Salmonella is believed to be the second most common source of food poisoning, and in the United States alone is estimated to be responsible for more than 1 million illnesses, over 19,000 hospitalizations, and more than 370 deaths every year.
(Cite CDC Food borne Illness Estimates)
In recent years we’ve seen outbreaks associated with Peanut butter, Papayas, sprouts, and currently, the CDC is investigation a multi-state outbreak linked to ground turkey.
This latest outbreak is of a strain known as Salmonella Heidelberg, which the CDC notes “is resistant to many commonly prescribed antibiotics; this antibiotic resistance can increase the risk of hospitalization or possible treatment failure in infected individuals.”
More than 2500 serotypes of Salmonella have been identified, and nearly all can cause illness in humans. For severe cases of severe Salmonellosis, antibiotics from the fluoroquinolone class - (ie. ciprofloxacin) are the drug of choice.
But since the 1990s, doctors have seen a steady rise in antibiotic resistance in a number of Salmonella strains. This from the World Health Organization.
Drug-resistant Salmonella
Multidrug-resistant (MDR) strains of Salmonella are now encountered frequently and the rates of multidrug-resistance have increased considerably in recent years. Even worse, some variants of Salmonella have developed multidrug-resistance as an integral part of the genetic material of the organism, and are therefore likely to retain their drug-resistant genes even when antimicrobial drugs are no longer used, a situation where other resistant strains would typically lose their resistance.
All of which serves as prelude to a new report in the Journal of Infectious Diseases on the rise, and global spread, of a multi-drug resistant strain of salmonella called S. Kentucky.
First a link the article, then I’ll return with more.
Simon Le Hello, Rene S. Hendriksen, Benoı ˆt Doublet, Ian Fisher, Eva Møller Nielsen, Jean M. Whichard,Brahim Bouchrif, Kayode Fashae, Sophie A. Granier, Nathalie Jourdan-Da Silva,Axel Cloeckaert, E. John Threlfall, Frederick J. Angulo, Frank M. Aarestrup, John Wain, and Francois-Xavier Weill
National Salmonella surveillance systems from France, England and Wales, Denmark, and the United States identified the recent emergence of multidrug-resistant isolates of Salmonella enterica serotype Kentucky displaying high-level resistance to ciprofloxacin.
Salmonella Kentucky was first isolated and described in the 1930s, by the Department of Animal Pathology in Lexington, Kentucky.
Although many of the news headlines this morning are portraying this resistant strain as something `new’, this strain has been spreading for nearly a decade.
in 2006, the lead author of today’s study published a letter in the CDC’s MMWR on the emergence of Ciprofloxacin resistant S. Kentucky among French travelers returning from northeast and eastern Africa in 2002.
Ciprofloxacin-resistant Salmonella Kentucky in Travelers
François-Xavier Weill,*
Sophie Bertrand,† Françoise Guesnier,* Sylvie Baucheron,‡ Patrick A.D. Grimont,* and Axel Cloeckaert‡
What is new is the number of drug-resistant cases, and their geographic spread, over the past few years. In 2002, there were 3 reported cases. In 2008, 174 cases were identified.
The resistant strain has been dubbed the ST198-X1 CIPR Kentucky clone to identify its resistant characteristics, and has been slowly increasing among isolates of Salmonella Kentucky identified in France, England and Wales, and Denmark over the past decade.
Specifically, these researchers found that:
- of the nearly 500 isolates of S. enterica serotype Kentucky tested in France, 200 were drug resistant.
- In England and Wales, roughly 1/3rd have been shown to be resistant. And in Demark, 45 of 114 isolates are listed as CIPR .
- Thus far, out of 679 S. Kentucky isolates identified in the United States, none have been found to be resistant.
This particular strain has been isolated from chickens and turkeys in several African nations (Ethiopia, Morocco, and Nigeria) and in the Middle East.
The authors suggest that the common use of fluoroquinolones - including ciprofloxacin - in chicken and turkey production in Nigeria and Morocco may have contributed to this strain’s emergence and spread.
All of which highlights the need for better surveillance of our global food supply, and for greater restrictions on the indiscriminate agricultural use of antibiotics.