#11,758
Although its primary findings were telegraphed 3 weeks ago (see Singapore MOH: 26 New Zika Cases & Preliminary Genetic Analysis) and again the following week (see Singapore MOH: Genetic Analysis Report On Local Zika Virus), we've a far more detailed report today in the Journal Eurosurveillance on the origins and evolution of Singapore's Zika outbreak.
As previously announced, the strain discovered in Singapore comes from an older branch of the Asian lineage, and was not imported from the Americas. Today's report also states:
Furthermore, in the tree, the long branch of the two new south-east Asian viruses found in this study suggests undetected evolution for several years.
Genetic analysis shows the Singapore strain to have evolved from branch that emerged before the Zika virus moved from the South Pacific to the Americas where it continues to evolve (see Emerging Microbes & Infect: Growing Genetic Diversity Of Zika Viruses In Latin America).
Eurosurveillance, Volume 21, Issue 38, 22 September 2016
Rapid communication
Citation style for this article: Maurer-Stroh S, Mak T, Ng Y, Phuah S, Huber RG, Marzinek JK, Holdbrook DA, Lee RT, Cui L, Lin RT. South-east Asian Zika virus strain linked to cluster of cases in Singapore, August 2016. Euro Surveill. 2016;21(38):pii=30347. DOI: http://dx.doi.org/10.2807/1560-7917.ES.2016.21.38.30347
Received:19 September 2016; Accepted:22 September 2016
Outbreak detection
On 22 August 2016, the Ministry of Health (MOH), Singapore, was informed by a general practitioner of a spate of cases presenting with non-specific symptoms, including rash, polyarthralgia and low grade fever. In addition, a number of cases had mild conjunctivitis. Four days later, one of the cases was referred to the Communicable Disease Centre, Tan Tock Seng Hospital. The patient’s blood and urine samples tested positive for Zika virus (ZIKV), and the blood was negative for both dengue and chikungunya viruses by polymerase chain reaction (PCR). Since the patient had no travel history within the past month, the MOH announced the first locally transmitted ZIKV infection on 27 August, after laboratory confirmation on a second set of blood and urine specimens. For the purpose of outbreak investigation and public health measures, MOH then directed the collection of clinical specimens from cases working or living in the surrounding areas, with fever (>37.8°C), rash, conjunctivitis and/or joint pain in the preceding two weeks. All clinical information and samples pertaining to the outbreak investigation were collected under the provisions of the Infectious Diseases Act in Singapore [1].
(SNIP)
Conclusions from sequence and structure analysis
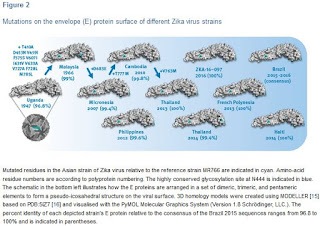
This study shows that there are still multiple ZIKV strains in circulation globally in 2016 which raises the question on their antigenic diversity or similarity for vaccine development. Therefore, we systematically mapped the outer surface envelope (E) protein changes from 1947 onwards across the whole Asian lineage (Figure 2).
At least within the Asian lineage the antigenic protein E surface is highly conserved and homogeneous with, from 2007 onwards, typical identities from 99.4% to 100% (or 3 to zero mutations) relative to the consensus of recent strains from Brazil. Moreover all of the E proteins in this lineage contain the typical N-glycosylation site (N444 in polyprotein numbering which is N154 in the E protein). The lack of surface mutation drift over the past 50 years (99% identity between Malaysia 1966 and consensus of Brazil 2015) also suggests that immune pressure on the E protein has not been a dominant factor in the Asian lineages’ virus fitness and evolution so far.
While the highly similar surface E proteins of the different strains in the Asian lineage should facilitate global vaccine development it is too early to judge if the strain linked to the Singapore outbreak would show any different disease characteristics from the one in French Polynesia and the Americas. Further studies are necessary and underway in Singapore to collect more data.