Credit CDC
#14,195
Just over three years ago the the CDC issued a Clinical Alert to U.S. Healthcare facilities about the Global Emergence of Invasive Infections Caused by the Multidrug-Resistant Yeast Candida auris.
C. auris is an emerging fungal pathogen that was first isolated in Japan in 2009. It was initially found in the discharge from a patient's external ear (hence the name `auris'). Retrospective analysis has traced this fungal infection back over 20 years.Since then we've been following the progress of this emerging pathogen - both in the United States and around the world - because:
Four weeks ago the CDC held an hour-long COCA (Clinician's Outreach & Communications Activity) call on this emerging threat called:
- C. auris infections have a high fatality rate
- The strain appears to be resistant to multiple classes of anti-fungals
- This strain is unusually persistent on fomites in healthcare environments.
- And it can be difficult for labs to differentiate it from other Candida strains
Multidrug-resistant Candida auris: Update on Current U.S. Epidemiology, Clinical Profile, Management, and Control StrategiesThe full video of this webinar presentation is posted at the above link, along with slides.
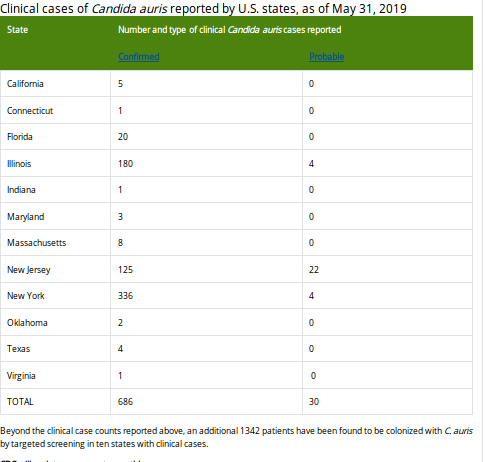
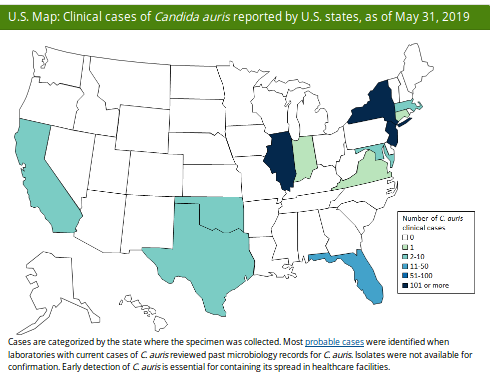
Last week the CDC published their June update on their C. Auris surveillance page, where they show - as of May 31th - 686 confirmed cases and 30 probable cases, across 12 states (primarily New York, New Jersey & Illinois).
Additionally, 1342 patients have been found to be asymptomatically colonized with C. auris through targeted screening conducted in six states with clinical cases.To put this into perspective, this represents a roughly 30% increase in confirmed cases since the EOY 2018 numbers (n=520), in just the first five months of 2019.
First some excerpts from the latest CDC update, followed by excerpts from a recently published study in the CDC's EID Journal.
While the numbers remain relatively small, they are undoubtedly under reported, both here in the United States, and around the world. In many places testing and surveillance is simply unavailable, while in other places, it is under under utilized.
Lastly, we have a new research letter, published this week in the CDC's EID Journal, detailing the finding of what appears to be a genetically distinct 5th clade of C. auris in Iran.
Research Letter
Potential Fifth Clade of Candida auris, Iran, 2018
Nancy A. Chow, Theun de Groot, Hamid Badali, Mahdi Abastabar, Tom M. Chiller, and Jacques F. Meis
Abstract
Four major clades of Candida auris have been described, and all infections have clustered in these 4 clades. We identified an isolate representative of a potential fifth clade, separated from the other clades by >200,000 single-nucleotide polymorphisms, in a patient in Iran who had never traveled outside the country.
In the last decade, Candida auris has emerged in healthcare facilities as a multidrug-resistant pathogen that can cause outbreaks of invasive infections (1). C. auris has now been identified in >35 countries, many of which have documented healthcare-associated person-to-person spread (2). Transmission of this yeast is facilitated by its ability to colonize skin and other body sites, as well as its ability to persist for weeks on surfaces and equipment (3).
Whole-genome sequencing of C. auris has identified 4 major populations in which isolates cluster by geography (4). These populations are commonly referred to as the South Asian (I), East Asian (II), African (III), and South American (IV) clades. Worldwide, C. auris isolates continue to cluster in 1 of the 4 clades (Figure; 5–7). We report an isolate representative of a fifth clade in Iran from a patient who never traveled outside that country. The patient was a 14-year-old girl in whom C. auris otomycosis had been diagnosed; her case was the first known C. auris case in Iran (8).
(SNIP)
The C. auris isolate from Iran appears to represent a fifth major clade. Although this case was reported in 2018, additional cases of C. auris infections and colonization are thought to exist in Iran, given that challenges in diagnostic capacity in the country have probably limited the identification of more C. auris cases. The patient in this case was reported to have never traveled outside Iran (8), suggesting that this population structure might not be a result of a recent C. auris introduction into the country and that it might have emerged in Iran some time ago. Determining whether additional C. auris cases exist in Iran and whether such strains are related will help shed light on how C. auris emerged in Iran.
The isolate from Iran was susceptible to the 3 major classes of antifungal drugs and was cultured from ear swab specimens from the patient (8). C. auris of the East Asian clade is thought to have a propensity for the ear that is uncharacteristic of the other major clades (9). A recent study showed that, of 61 C. auris isolates obtained from 13 hospitals across South Korea during a 20-year period, 57 (93%) came from ear cultures (10). Although a systematic analysis has not been conducted, there are limited reports of ear infections or colonization caused by C. auris of the South Asian, African, or South American clades, so it is of interest that the isolate from Iran was most closely related to isolates of the East Asian clade, albeit with a difference of hundreds of thousands of single nucleotide polymorphisms. Ultimately, our discovery is a reminder that much about C. auris remains to be learned and underscores the need for vigilance in areas where C. auris has not yet emerged.
Dr. Chow is a molecular epidemiologist in the Mycotic Diseases Branch of the Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention. Her primary research interests include application of whole-genome sequencing and metagenomics for outbreak investigations as well as integrating and visualizing epidemiologic and laboratory data sources.