#14,172
Yesterday, in
Viruses: A Global Perspective on H9N2 Avian Influenza Virus, we looked at an excellent review of the evolution and geographic spread of various strains and lineages of the LPAI H9N2 virus over the past two decades.
Although a low path (LPAI) virus in poultry,
H9N2 continues to show worrying signs of mammalian adaptation and has lent its genes (via reassortment) to a number of other viruses (see The Lancet: H9N2’s Role In Evolution Of Novel Avian Influenzas),
While the incidence of H5N1, H5N6, and H7N9 infections has plummeted in China the past two years, recently we've seen increases in H9N2 outbreaks across
Africa,
Asia, and the
Middle East, and starting in 2015, a substantial jump in the number of human infections reported (see
FluTrackers List).
With China's massive H5+H7 poultry vaccination program -
at least temporarily - reducing the risks of a novel flu pandemic emerging from that region, our eyes turn to areas of the world where H5Nx and H9N2 viruses continue to co-circulate, and where surveillance and containment is poor.
And the Middle East - where HPAI H5N1, HPAI H5N8, and HPAI H5N2 all circulate along side LPAI H9N2 - fits that bill.
While no naturally occurring
H9N2/H5Nx reassortants have been reported in the Middle East, five months ago, in
J. Virology: Genetic Compatibility of Reassortants Between Avian H5N1 & H9N2 Influenza Viruses, we saw a study of lab generated reassortments of Egyptian H5N1 and H9N2 that found:
". . . our analyses indicated a
substantial emergence potential of influenza virus reassortants derived
from the H5N1 and H9N2 viruses currently cocirculating in Egypt, as well
as the possibility of their high public health risk for humans relative
to the parental H5N1 and H9N2 viruses. Cocirculation of the two
influenza virus subtypes in birds may accelerate the emergence of novel
viruses that may be a public health risk."
These same researchers are back with a new research article in
PloS Pathogens, which looks at some of the PB2 mutations in LPAI H9N2 viruses circulating in the Middle East and discusses the potential for reassortant viruses to emerge.
This is a lengthy and detailed open access study, and you'll want to follow the link to read it in its entirety. I'll have a brief postscript when you return.
Research Article
PB2 mutations arising during H9N2 influenza evolution in the Middle East confer enhanced replication and growth in mammals
Yasuha Arai,Norihito Kawashita, Madiha Salah Ibrahim, Emad Mohamed Elgendy, Tomo Daidoji, Takao Ono, Tatsuya Takagi, Takaaki Nakaya, Kazuhiko Matsumoto, Yohei Watanabe
Published: July 2, 2019
https://doi.org/10.1371/journal.ppat.1007919
Abstract
Avian influenza virus H9N2 has been endemic in birds in the Middle East, in particular in Egypt with multiple cases of human infections since 1998. Despite concerns about the pandemic threat posed by H9N2, little is known about the biological properties of H9N2 in this epicentre of infection.
Here, we investigated the evolutionary dynamics of H9N2 in the Middle East and identified phylogeny-associated PB2 mutations that acted cooperatively to increase H9N2 replication/transcription in human cells. The accumulation of PB2 mutations also correlated with an increase in H9N2 virus growth in the upper and lower airways of mice and in virulence.
These mutations clustered on a solvent-exposed region in the PB2-627 domain in proximity to potential interfaces with host factors. These PB2 mutations have been found at high prevalence during evolution of H9N2 in the field, indicating that they have provided a selective advantage for viral adaptation to infect poultry.
Therefore, continuous prevalence of H9N2 virus in the Middle East has generated a far more fit or optimized replication phenotype, leading to an expanded viral host range, including to mammals, which may pose public health risks beyond the current outbreaks.
Author summary
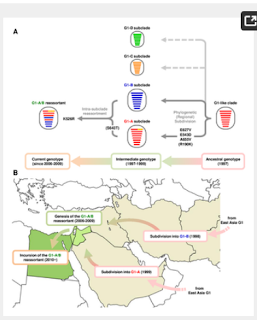
The G1-like clade of H9N2 influenza viruses can undergo genetic reassortment with other influenza virus subtypes to produce novel zoonotic viruses, such as the Gs/GD lineage H5N1, H7N9, H10N8, and H5N8 viruses.
Since 1998, the G1-like subclade of H9N2 influenza virus has been widely circulating in birds in Central Asia and the Middle East and a number of human cases have been reported. However, little is known about the biological properties of H9N2 viruses in this epicentre of infection.
Our data showed that, during about two decades of evolution in nature, G1-like subclade strains evolved to produce strains with appreciably higher replication phenotypes in Central Asia and the Middle East, which led to their expanded host range, including to humans.
Therefore, G1-like subclade strains in these areas may accumulate mutations to produce novel viruses and the large gene pool in these areas would enable reassortment with other influenza viruses. This study indicated the need for studies of H9N2 viruses in such areas to monitor their evolutionary dynamics and possible genetic changes.
(SNIP)
The H9N2 virus and the H5N1 clade 2.2.1 viruses have been prevalent in poultry in Egypt, which has provided the opportunity for reassortment between the two viruses. Fortunately, the emergence of H9N2/H5N1 reassortants has not been reported in Egypt thus far.
However, we recently reported that Egyptian H9N2 and H5N1 viruses have a high genetic compatibility and a substantial potential to generate reassortants with greater fitness in mammalian species than the parental viruses [20]. The results of this study suggested that, if an H5N1 clade 2.2.1 virus successfully recruited the G1-A/B PB2 gene, such reassortants may pose an infection threat beyond the current outbreaks in the Middle East.
The G1-A/B reassortant has an increased replication ability in human cells at 33°C and in the upper respiratory tract of mice. This implied that an exposure to fewer G1-A/B reassortant viruses may establish an infection in the respiratory tract of a mammalian species, greatly increasing the likelihood of a co-infection and reassortment with a seasonal human or swine influenza virus in the upper airway. The G1-A/B reassortant has an HA-Q226L mutation (H3 numbering) that increased viral binding affinity to human respiratory epithelia [42]. In view of this concern, it should be noted that seroepidemiological studies have indicated substantial exposure of humans and pigs to both H5N1 and H9N2 viruses in Egypt [43, 44].
A high level of genetic compatibility between H9N2 and seasonal H1N1 2009 viruses also has been reported [45, 46].
Taken together, these findings suggested that the Middle East, with an epicenter in Egypt, is now a hot spot of H9N2 and H5N1 virus evolution, which may generate novel viruses presenting an increased public health risk.
In conclusion, this study showed that the influenza virus G1-A/B reassortant, that has been prevalent in Egypt, has diversified phylogenetically in the Middle East during its natural evolution and generated a distinct phenotype with an expanded host range to mammals, including humans. Our results highlighted the need for studies of H9N2 viruses to trace their evolutionary dynamics in this area and closely monitor possible genetic changes in these viruses. In addition, implementation of effective control measures may be needed to reduce the risk of a future pandemic.
(Continue . . . )
While a standalone H9N2 virus may not be at the top of our pandemic threats list, it is regarded as having at least
some pandemic potential by our CDC (see
IRAT SCORE), and
several candidate vaccines have been developed.
The bigger risk comes from its promiscuous nature and ability to reassort with other, potentially more dangerous, flu viruses.
The three avian flu viruses of greatest concern over the past decade –
H5N1, H7N9, and
H5N6 – all share a couple of important features (see
Study: Sequence & Phylogenetic Analysis Of Emerging H9N2 influenza Viruses In China):
- They have all come about through viral reassortment in poultry
- And while their HA and NA genes may differ - they all carry the internal genes from the avian H9N2 virus
While the next novel pandemic virus could emerge anywhere, the rapid evolution of H9N2 in the Middle East - where at least 3 different HPAI H5 viruses also circulate -
appears to have raised the stakes in that part of the world.
And it is, sadly, another region of the world where surveillance, and our visibility, are less than optimal.