#16,604
Although incredibly well-adapted to humans, the SARS-CoV-2 virus is also known to infect more than 2 dozen other mammalian species, and has shown signs of increasing its host range as it evolves. The big concern is this virus may take divergent evolutionary paths in these other hosts, and produce variants that could `spill back' into humans down the road.
And in fact, we've already seen that happen with farmed mink.
In November of 2020 SARS-CoV-2 jumped from humans to mink in Denmark, spread like wildfire, and began to mutate into new mink-variants (see Denmark Orders Culling Of All Mink Following Discovery Of Mutated Coronavirus).
Several mutated viruses jumped back into humans, and began to spread in the community (see WHO 2nd Update: SARS-CoV-2 mink-associated variant strain – Denmark), forcing North Denmark To Lockdown Over Mutated Coronavirus Concerns.
This emergency was relatively short-lived, as the Alpha variant emerged in Europe in late 2020 and quickly supplanted these mink-variants. But it did demonstrate the problem; carriage of SARS-CoV-2 by other host species can produce new variants, which can jump back into humans.
Since then we've seen mink farms around the world (including in the United States) infected with COVID, and additional evidence that humans have been infected by mink-variant viruses (see CDC: Investigating Possible Mink-To-Human Transmission Of SARS-CoV-2 In The United States).
We saw a similar scenario play out in Hong Kong (see Hong Kong Detects COVID In Pet Store Hamsters - Suspends Sales & Orders Cull) in early January (see Nature How sneezing hamsters sparked a COVID outbreak in Hong Kong).
Fortunately, most farmed animals (pigs, chickens, cattle, etc.) are poor hosts for the SARS-CoV-2 virus. Dogs and cats are mildly susceptible, but since they don't have contact with hundreds of other animals, aren't as likely to generate mutations.
There are concerns that SARS-CoV-2 may be spreading silently in other wildlife - such as rodents and semi-aquatic mammals - but biggest findings thus far have been in North American White-Tailed Deer (WTD). A few of many studies we've seen since last summer include:
Preprint: Evolutionary Trajectories of SARS-CoV-2 Alpha and Delta Variants in White-Tailed Deer in Pennsylvania
More Documented Spillovers Of COVID-19 Into North American Deer
Nature: SARS-CoV-2 Infection in Free-ranging White-tailed Deer
Two New Reports Find Widespread SARS-CoV-2 In North American Deer
While this growing reservoir of SARS-CoV-2 in North American deer has been a growing concern, until very recently the viruses that have been detected have been quite similar to the variants currently (or previously) circulating in humans.
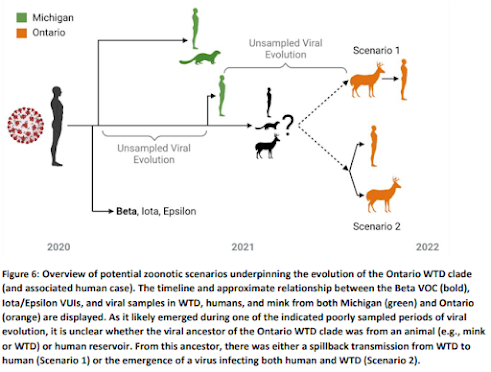
And today, we have a preprint from researchers in Ontario Canada that finds Highly divergent SARS-CoV-2 variants in WTD, and potential evidence of deer-to-human transmission. This 31-page page is highly detailed, and while there are admittedly gaps in the data, makes a plausible case for a spillback of a WTD-Variant into humans (see graphic below).But last week's report on Evolutionary Trajectories of SARS-CoV-2 reported finding ". . .deer-derived alpha variants diverged significantly from those in humans, consistent with a distinctive evolutionary trajectory in deer."
Highly divergent white-tailed deer SARS-CoV-2 with potential deer-to-human transmission
Brad Pickering, Oliver Lung, Finlay Maguire, Peter Kruczkiewicz, Jonathon D Kotwa, Tore Buchanan, Marianne Gagnier, Jennifer Guthrie, Claire Jardine, Alex Marchand-Austin, Ariane Masse, Heather McClinchey, Kuganya Nirmalarajah, Patryk Aftanas, Juliette Blais-Savoie, Hsien-Yao Chee, Emily Chien, Winfield Yim, Melissa Goolia, Matthew Suderman, Mathieu Pinette, Greg Smith, Daniel Sullivan, Jossip Rudar, Elizabeth Adey, Michelle Nebroski,Marceline Cote, Genevieve Laroche, Allison McGeer, Larissa Nituch, Samira Mubareka, Jeff Bowman
doi: https://doi.org/10.1101/2022.02.22.481551
Abstract
Wildlife reservoirs of SARS-CoV-2 can lead to viral adaptation and spillback from wildlife to humans (Oude Munnink et al., 2021). In North America, there is evidence of spillover of SARS-CoV-2 from humans to white-tailed deer (Odocoileus virginianus), but no evidence of transmission from deer to humans (Hale et al., 2021; Kotwa et al., 2022; Kuchipudi et al., 2021).
Through a multidisciplinary research collaboration for SARS-CoV-2 surveillance in Canadian wildlife, we identified a new and highly divergent lineage of SARS-CoV-2. This lineage has 76 consensus mutations including 37 previously associated with non-human animal hosts, 23 of which were not previously reported in deer. There were also mutational signatures of host adaptation under neutral selection.
Through a multidisciplinary research collaboration for SARS-CoV-2 surveillance in Canadian wildlife, we identified a new and highly divergent lineage of SARS-CoV-2. This lineage has 76 consensus mutations including 37 previously associated with non-human animal hosts, 23 of which were not previously reported in deer. There were also mutational signatures of host adaptation under neutral selection.
Phylogenetic analysis revealed an epidemiologically linked human case from the same geographic region and sampling period. Together, our findings represent the first evidence of a highly divergent lineage of SARS-CoV-2 in white-tailed deer and of deer-to-human transmission.
(SNIP)
Potential deer-to-human transmission
Our phylogenetic analysis also identified a human-derived sequence from Ontario (ON-PHL-21-44225) that was both highly similar (80/90 shared mutations; Table S2) and formed a well-supported monophyletic group (100% UFB) with the WTD samples (Figure 3). The small number of samples and relative diversity within the WTD clade make it difficult to determine the exact relationship between the human sample and other WTD samples (78% UFB for a most recent common ancestor with 4658). However, global (FIgure 2) and focal (Figure 3) ML analyses and an Usher-based (Turakhia et al., 2021) (Figure S6) parsimony analysis all support this human sample belonging to the WTD clade.
The human sequence also has a plausible epidemiological link to the WTD samples as it was collected in the same geographical region (Southwestern Ontario), during the same time period (autumn 2021) after having known close contact with deer. At the time of the human case detection 100% of eligible confirmed PCR positive SARS-CoV-2 samples collected from human cases were requested by Public Health Ontario and Ontario COVID-19 Genomic Network partners for genome sequencing, and no other genetically related human-derived samples were identified. However, it should be noted that not all requested samples are received and/or successfully sequenced, and the surge of Omicron cases necessitated a reduction in the proportion human-derived SARS-CoV-2 sampled for sequencing in the region, moving from 100% to 50%, 20%, and 5% sampling on the 7th, 20th, and 30th of December 2021 respectively (Public Health Ontario, 2022)
(SNIP)
At this time, there is no evidence of recurrent deer to human or sustained human to human transmission of the Ontario WTD SARS-CoV-2 clade. However, the emergence of Omicron and the end of deer hunting season has meant both human and WTD testing and genomic surveillance in this region has been limited since these samples were collected. Therefore, we cannot determine with certainty whether the lack of additional human cases reflects no onward transmission from the human case, no further spillover events from WTD, or limited genomic surveillance. Enhanced surveillance is of particular importance given human population density and mobility in the region, coupled with WTD population dynamics.
This work underscores the need for a broader international One Health lens to identify new intermediate or reservoir hosts capable of driving sustained transmission and divergent viral evolution. Selective advantage has led to the emergence of new variants outcompeting those in circulation. However, the absence of a variant in the human population does not mean universal absence of variants across a much broader range of under-sampled potential host species. To date, many sampling strategies have been based on access and convenience. Focusing efforts at human-animal interfaces and integration of human epidemiological data would enable analysis of determinants of spillover and inter-species transmission. A broader analysis examining human drivers of spillover and spillback and knock-on effects on wildlife and human health is urgently needed to identify, develop and implement mitigation strategies, beginning with reducing viral activity in humans.
However, these are just the tip of the iceberg as the susceptibility of most terrestrial wild animals to SARS-CoV-2 has not been tested. In addition, the research on susceptibility of marine wildlife (especially marine mammals) to SARS-CoV-2 is still lacking. Due to frequent marine human activities (such as mariculture and marine fishing), the frequency of human contact with marine organisms is high. If some marine organisms are highly susceptible to SAR-CoV-2, there is a risk that SARS-CoV-2 could be transmitted from humans to marine organisms, and worse, SARS-CoV-2 then might spread in the marine ecosystem, which may lead to the generation of some novel SARS-CoV-2 variants with unknown threats to humans.
Therefore, it is necessary to carry out large-scale SARS-CoV-2 screening for terrestrial and marine wildlife, especially those susceptible ones, in order to monitor the status of infection and mutation of SARS-CoV-2 in wild animals, so as to formulate further prevention and control strategies. It also provides more clues to the study of the origin and cross-species transmission of SARS-CoV-2.
While the co-circulation of SARS-CoV-2 in other mammalian species doesn't guarantee we'll be faced with the emergence of new, and possibly more dangerous, variants in the future - it would appear to raise the risks.
We've two examples (Denmark and Hong Kong) of SARS-CoV-2 spillback from animal hosts, and there are some researchers who believe Omicron may have evolved in mice, and then spilled back as well.
While our pandemic-weary world seems intent on declaring victory and moving on, we need to consider that the virus may have other plans.