#17,489
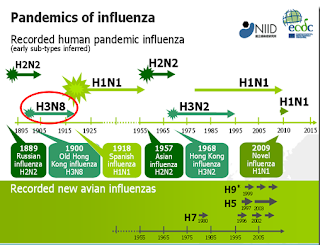
Although our knowledge of pandemic influenza viruses only goes back about 130 years - and anything before 1918 is fairly shaky - as near as we can tell, all human influenza pandemics have come from H1, H2, or H3 viruses (see Are Influenza Pandemic Viruses Members Of An Exclusive Club?).
Novel H1, H2, and H3 flu viruses - even though they also have an avian origin - appear to have fewer barriers to overcome in order to jump to humans. They may not prove to be as virulent as H5 & H7 avian subtypes, but that puts them at or near the top of our pandemic threats list.While H5N1 currently sits atop our pandemic worry list, no one really knows if a non-H1/H2/H3 virus can cause a human pandemic. But just because one hasn't (recently), doesn't mean it can't.
EID Journal: Zoonotic Threat of G4 Genotype Eurasian Avian-Like Swine Influenza A(H1N1) Viruses, China, 2020
EID Dispatch: Replication of Novel Zoonotic-Like Influenza A(H3N8) Virus in Ex Vivo Human Bronchus and Lung
China: Emergence of a Novel Reassortant H3N6 Canine Influenza Virus
Increased Public Health Threat of Avian-origin H3N2 Influenza Virus During Evolution in Dogs (Revisited)
While these are the only 3 confirmed human infections with H3N8, we've been following its evolution in birds, dogs, horses, and marine mammals for quite some time.
- H3N8 remains a plausible cause of a global influenza pandemic that spread out of Russia in 1889-1900 (although some researchers now suspect a coronavirus instead).
- about 60 years ago H3N8 jumped unexpectedly to horses, supplanting the old equine H7N7 and is now the only equine-specific influenza circulating the globe
- in 2004 the equine H3N8 virus mutated enough to jump to canines, and began to spread among greyhounds in Florida (see EID Journal article Influenza A Virus (H3N8) in Dogs with Respiratory Disease, Florida).
- in 2011 avian H3N8 was found in marine mammals (harbor seals), and 2012’s mBio: A Mammalian Adapted H3N8 In Seals, provided evidence that this virus had recently adapted to bind to alpha 2,6 receptor cells, the type found in the human upper respiratory tract.
- in 2015's J.Virol.: Experimental Infectivity Of H3N8 In Swine, we saw a study that found that avian (but not canine or equine) H3N8 could easily infect pigs.
- And just last March, in Emerging Microbes & Inf.: Prevalence, Evolution, Replication and Transmission of H3N8 Avian Influenza Viruses, we looked at the rapid spread of H3N8 (and other) avian flu viruses in Eastern China
Authors: Peiwen Chen, Ziying Jin, Liuxia Peng, Zuoyi Zheng, Yiu-Man Cheung, Jing Guan, Liming Chen, SHOW ALL (24 AUTHORS), Yi Guan
ABSTRACT
Although influenza A viruses of several subtypes have occasionally infected humans, to date only those of the H1, H2, and H3 subtypes have led to pandemics and become established in humans. The detection of two human infections by avian H3N8 viruses in April and May of 2022 raised pandemic concerns. Recent studies have shown the H3N8 viruses were introduced into humans from poultry, although their genesis, prevalence, and transmissibility in mammals have not been fully elucidated.Findings generated from our systematic influenza surveillance showed that this H3N8 influenza virus was first detected in chickens in July 2021 and then disseminated and became established in chickens over wider regions of China. Phylogenetic analyses revealed that the H3 HA and N8 NA were derived from avian viruses prevalent in domestic ducks in the Guangxi-Guangdong region, while all internal genes were from enzootic poultry H9N2 viruses. The novel H3N8 viruses form independent lineages in the glycoprotein gene trees, but their internal genes are mixed with those of H9N2 viruses, indicating continuous gene exchange among these viruses.Experimental infection of ferrets with three chicken H3N8 viruses showed transmission through direct contact and inefficient transmission by airborne exposure. Examination of contemporary human sera detected only very limited antibody cross-reaction to these viruses. The continuing evolution of these viruses in poultry could pose an ongoing pandemic threat.
IMPORTANCEA novel H3N8 virus with demonstrated zoonotic potential has emerged and disseminated in chickens in China. It was generated by reassortment between avian H3 and N8 virus(es) and long-term enzootic H9N2 viruses present in southern China. This H3N8 virus has maintained independent H3 and N8 gene lineages but continues to exchange internal genes with other H9N2 viruses to form novel variants. Our experimental studies showed that these H3N8 viruses were transmissible in ferrets, and serological data suggest that the human population lacks effective immunological protection against it. With its wide geographical distribution and continuing evolution in chickens, other spillovers to humans can be expected and might lead to more efficient transmission in humans.
(SNIP)DISCUSSIONThis study aimed to investigate the genesis and emergence of a recently identified zoonotic H3N8 virus. Based on the phylogenetically closest viruses, our surveillance data showed that the H3N8 virus most likely originated in the Guangxi-Guangdong region of China. We were also able to show that the H3N8 viruses had been circulating in chickens for over a year at a high prevalence and had disseminated to at least seven provinces before detection in humans (Fig. 1). We detected an obvious seasonality that was also observed in the H7N9/2013 influenza viruses (10). Like the H7N9/2013 and H10N8/2013 viruses, the H3N8 viruses mixed their duck-origin HA and NA genes with internal genes from chicken H9N2 viruses enzootic in the region (5, 8, 10, 11). This demonstrates again that the influenza ecosystem of China, with its high and frequently interacting human and poultry populations, is a major source of novel influenza viruses with zoonotic capacity.
(SNIP)H3 viruses are one of only three influenza subtypes to have caused a pandemic, and the H3 subtype has shown the greatest propensity to infect mammals (18, 21, 22). Apart from sporadic infections in aquatic mammals (19) and carnivores (26), this subtype has become established in swine, equids, and canids (18, 27, 28). Of the persistent mammalian H3 lineages, three are of whole or partial avian origin, with the human and canine H3N2 lineages emerging in China (18). As H3 is a major influenza subtype in the wild birds in China, monitoring and control of H3 viruses entering the agricultural or human environments seem advisable.
As we've discussed often, Southeast Asia has long been considered `the cradle of influenza’; an area of the world where both human and animal influenza viruses circulate more-or-less year round, and where there are ample opportunities for viruses to spillover (both to, and from, animals).
Although the 2009 H1N1 virus appears to have emerged from swine in Mexico, and the 1918 H1N1 may have originated from the United States, China has been the springboard for H2N2 (in 1957), H3N2 ( in 1968), and more recently H5N1, H5N6, H7N9, H10N3, H3N6 and H3N8 (to name a few).
Surveillance and reporting from Mainland China (and other parts of Southeast Asia) are often lacking - and are frequently held close to the vest for political and economic reasons - meaning we could be easily be blindsided by the next emerging threat.
An H5 or H7 influenza pandemic might have a higher case fatality rate (CFR), but history suggests that H1, H2, or H3 pandemics are more likely to happen.
While hopefully less deadly, COVID has demonstrated that even a 1% CFR pandemic can have devastating impacts on our modern world.
