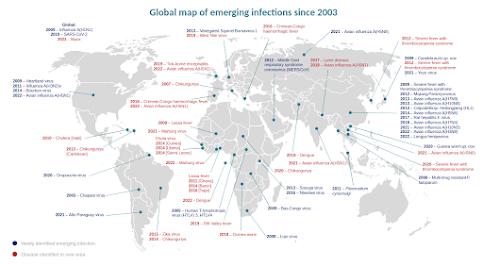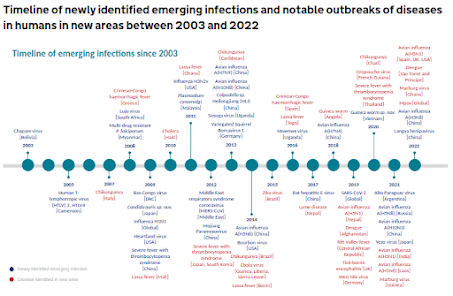
Credit UK HAIRS
#17,769
A little over a decade ago, in mBio: A Strategy To Estimate The Number Of Undiscovered Viruses, we saw an estimate that there were at least 320,000 unidentified mammalian viruses awaiting discovery. Even if we dropped a zero or two, that's still an impressive reservoir of potential disease threats flying under our radar.
While most will never pose a threat to public health, some percentage have - or will eventually develop - zoonotic potential.
The UK HAIRS chart above shows that over the past two decades, the number of new zoonotic infections emerging each year - and the global spread of already known threats - have both increased markedly.
Although new zoonotic threats can emerge anywhere in the world (see HAIRS map below), Asia - and China in particular - have been source for many (see EID Journal on Predicting Hotspots for Influenza Virus Reassortment).
Last month we looked at V. Sinica: Diversity and Independent Evolutionary Profiling of Rodent-borne Viruses in Hainan, China, where researchers described the discovery of 8 previously unknown pathogens with zoonotic potential circulating in rodents on Hainan Island, China.
Today, in a similar vein, we have another research paper reports on the diversity and ecological complexity of bat CoVs in southern China. Testing anal swaps gathered from 729 bats over a 2 year period (2016-2017), researchers detected 58 bat CoVs from 9 different species, and 6 locations.
This is a lengthy, and detailed report, so I've only posted the abstract and a few brief excerpts. Follow the link to read it in its entirety. I'll have a postscript after the break.
Pangolin HKU4-related coronaviruses found in greater bamboo bat from southern ChinaMin Guo a 1, Kai Zhao b 1, Xingwen Peng a, Xiangyang He a, Jin Deng a, Bo Wang c, Xinglou Yang b d, Libiao Zhang a
Under a Creative Commons license open access
Highlights
• Nine bat species from Yunnan and Guangdong provinces of China carried 58 bat CoVs.• Two representative full-length genomes of bat CoVs, TyRo-CoV-162275 and TyRo-CoV-162269, were obtained.
• TyRo-CoV-162275 genome had the highest identity with Malayan pangolin HKU4-related coronaviruses.
• TyRo-CoV-162275 found in greater bamboo bat is the first bat HKU4r-CoV reported with furin protease cleavage site.
Abstract
Coronavirus (CoV) spillover originating from game animals, particularly pangolins, is currently a significant concern. Meanwhile, vigilance is urgently needed for coronaviruses carried by bats, recognised as natural reservoirs of many coronaviruses.
In this study, we collected 729 anal swabs of 20 different bat species from nine locations in Yunnan and Guangdong provinces, southern China, in 2016 and 2017 and described the molecular characteristics and genetic diversity of alphacoronaviruses (αCoVs) and betacoronaviruses (βCoVs) in bats.
Using RT-PCR, we detected 58 (8.0%) bat CoVs in nine bat species from six locations. Furthermore, using the Illumina platform, we sequenced two representative full-length genomes of the bat CoVs, TyRo-CoV-162275 and TyRo-CoV-162269.
Sequence analysis showed that TyRo-CoV-162275 shared the highest identity with Malayan pangolin (Manis javanica) HKU4-related coronaviruses (MjHKU4r-CoVs) from Guangxi Province, whereas TyRo-CoV-162269 was closely related to HKU33-CoV discovered in a greater bamboo bat (Tylonycteris robustula) from Guizhou Province.
Notably, TyRo-CoV-162275 has putative furin protease cleavage site in its S protein and likely uses human dipeptidyl peptidase-4 (hDPP4) as a cell-entry receptor, similar to MERS-CoV.
To the best of our knowledge, this is the first report of a bat HKU4r-CoV strain with a furin protease cleavage site. These findings expand our understanding of coronavirus geographic and host distributions.
(SNIP)
4. Discussion
Bats play important roles in the emergence of zoonotic viruses. Most bat-borne viruses have been discovered through passive origination studies during disease outbreak investigations. During the past decades, several coronavirus disease outbreaks, such as SARS-CoV in 2002–2003, MERS-CoV in 2012, and SARS-CoV-2 in 2019, had severe impacts on human health and the world economy.
To date, approximately 5,000 bat coronavirus sequences have been reported, and bats are recognized as the major natural reservoirs of alpha- and beta-coronaviruses. Recent studies have shown that approximately 66,280 people are infected with SARSr-CoVs annually in Southeast Asia (Sanchez et al., 2022).
We are currently in the era of proactive coronavirus discovery to fight future bat coronaviruses. We conducted a bat coronavirus survey in southern China, which has a wide distribution and rich diversity of bats. Fifty-eight coronavirus sequences have been identified, including HKU10r-CoV, HKU6r-CoV, SARSr-CoV, HKU2r-CoV, and MERSr-CoV. Most importantly, a cluster of HKU4r-CoVs was detected in greater bamboo bats, providing new evidence that pangolins are the HKU4r-CoVs intermediate reservoir (Chen et al., 2023).
(SNIP)
5. Conclusions
In conclusion, our findings demonstrate the greater diversity and ecological complexity of bat CoVs in southern China, Yunnan and Guangdong provinces than previously appreciated. We found that pangolin HKU4-related coronaviruses in T. robustula contain a furin protease cleavage site. Further studies and characterisations of bat CoVs should be conducted to provide additional insights into their host range and the evolutionary history of bat populations in China and Southeast Asia.
While the origins of COVID remain hotly debated, what is abundantly clear is we've seen a remarkable number of coronaviruses detected in bats and other mammals around the world. A short list includes:
J. Med. Virology: Potential Cross-Species Transmission Risks of Emerging Swine Enteric Coronavirus to Human BeingsBefore SARS-CoV-2 burst onto the scene in early 2020, MERS-CoV (first reported in 2012) was the coronavirus thought to have the greatest pandemic potential (see 2017's A Pandemic Risk Assessment Of MERS-CoV In Saudi Arabia).
Nature: Virus Diversity, Wildlife-Domestic Animal Circulation and Potential Zoonotic Viruses of Small Mammals, Pangolins and Zoo Animals
Nature: Fatal Swine Acute Diarrhoea Syndrome Caused By An HKU2-related Coronavirus Of Bat Origin
Emerg. Microbes & Infect.: Novel Coronaviruses In Least Horseshoe Bats In Southwestern China
PNAS: SARS-like WIV1-CoV Poised For Human Emergence
While we don't hear as much about MERS-CoV as we used to, it still circulates in camels in the Middle East and in parts of Africa - occasionally spills over into humans - and remains a legitimate threat.
Two weeks ago, in BMJ Global: Historical Trends Demonstrate a Pattern of Increasingly Frequent & Severe Zoonotic Spillover Events, we looked at as study that examined the rise of zoonotic disease spillover events (and resultant deaths) since the early 1960s, and concludes that their rate has steadily increased over time.
Should those trends continue, they expect `. . . these pathogens to cause four times the number of spillover events and 12 times the number of deaths in 2050, compared with 2020.'
Coronaviruses are only part of the picture, of course. They are joined by novel influenza viruses, hemorrhagic fevers, henipaviruses, arboviruses, and many others which have zoonotic potential.
While we might go years, or even decades, before the next pandemic strikes there are no guarantees it won't kick off tomorrow.
Which is why we need to be using this time wisely, preparing for whatever nature throws at us next.