Credit Wikipedia
#17,736
In the wake of the surprise SARS-COV epidemic of 2002-2003 (see SARS And Remembrance) researchers around the world began a concerted search for new potential pandemic threats among coronaviruses in the wild.
- Two - MERS-CoV in the Middle East, and SARS-CoV-2 in China - would emerge in humans without warning.
- Many others, like 2016's PNAS: SARS-like WIV1-CoV Poised For Human Emergence, and 2017's EID Journal: A New Bat-HKU2–like Coronavirus in Swine, China, 2017, would be picked up by enhanced surveillance.
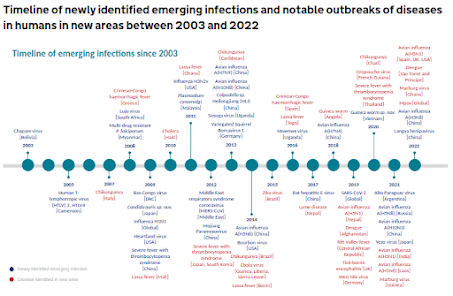
A little over a decade ago, in mBio: A Strategy To Estimate The Number Of Undiscovered Viruses, we saw an estimate that there were at least 320,000 unidentified mammalian viruses awaiting discovery.
While most are unlikely to pose a threat to public health, some will have zoonotic potential. A few recent examples include:
- Last year, in J. Virus Erad: A Review Of The Langya Virus Outbreak in China, 2022, a previously unknown henipavirus was detected in 35 humans in China.
- A year before that, researchers in Australia identified A Novel Variant Of the Hendra Virus
- And earlier this year we saw a preprint describing a Bat-borne Influenza A Virus H9N2 Exhibits a Set of Unexpected Pre-pandemic Features.
More than 680 swabs were collected from rodents between 2017 and 2021, and among their discoveries was a novel coronavirus dubbed CoV-HMU-1. Other novel viruses discovered included two new pestiviruses , two new parvoviruses , two new papillomaviruses, and a new astrovirus.
Due to its length I've only posted the Abstract and a small snippet from the Discussion/Conclusion. Those wishing a deeper dive will want to follow the link to read it in its entirety.
I'll have a brief postscript after the break.
Diversity and independent evolutionary profiling of rodent-borne viruses in Hainan, a tropical island of China
Youyou Li c d e 1, Chuanning Tang c d e 1, Yun Zhang c d e 1, Zihan Li c d e 1, Gaoyu Wang c d e, Ruoyan Peng c d e, Yi Huang c d e, Xiaoyuan Hu c d e, Henan Xin a, Boxuan Feng a, Xuefang Cao a, Yongpeng He a, Tonglei Guo a, Yijun He a, Haoxiang Su a, Xiuji Cui c d e, Lina Niu c d e, Zhiqiang Wu a, Jian Yang a, Fan Yang a…Jiang Du a c d
https://doi.org/10.1016/j.virs.2023.08.003Get rights and content
Under a Creative Commons license
Highlights
• The composition of the rodent-borne virome in Hainan Province, China was analyzed.
• Multiple novel viruses carried by rodents in the Hainan Province were identified.
• Potential zoonosis viruses have evolved independently in inaccessible areas.
Abstract
The risk of emerging infectious diseases (EID) is increasing globally. More than 60% of EIDs worldwide are caused by animal-borne pathogens. This study aimed to characterize the virome, analyze the phylogenetic evolution, and determine the diversity of rodent-borne viruses in Hainan Province, China.
We collected 682 anal and throat samples from rodents, combined them into 28 pools according to their species and location, and processed them for next-generation sequencing and bioinformatics analysis. The diverse viral contigs closely related to mammals were assigned to 22 viral families.
Molecular clues of the important rodent-borne viruses were further identified by polymerase chain reaction for phylogenetic analysis and annotation of genetic characteristics such as arenavirus, coronavirus, astrovirus, pestivirus, parvovirus, and papillomavirus.
We identified pestivirus and bocavirus in Leopoldoms edwardsi from Huangjinjiaoling, and bocavirus in Rattus andamanensis from the national nature reserves of Bangxi with low amino acid identity to known pathogens are proposed as the novel species, and their rodent hosts have not been previously reported to carry these viruses.
These results expand our knowledge of viral classification and host range and suggest that there are highly diverse, undiscovered viruses that have evolved independently in their unique wildlife hosts in inaccessible areas.
(SNIP)
Overall, the virome reported in this study is highly diverse. These viruses may be species-specific and distribute among these endemic hosts before spilling over. These results extend our knowledge of viral taxonomy and host range, and show that, in inaccessible areas, there are still highly diverse viruses that have evolved independently in their unique wildlife hosts. If these viruses cross the host barrier, they are highly possible to cause zoonosis.
5. Conclusions
Our results provide a profile of the composition of the rodent-borne virus and a baseline for rodent-borne viruses in Hainan Province, China. We detected PestVs and bocaviruses with low identities that were not previously reported to be carried by rodents. The associated viral genome sequences obtained in our study were corroborated, allowing us to further understand the phylogenetic relationship of the viruses in Hainan Province to other regions and obtain data on the diversity and independent evolutionary profiling of rodent-borne pathogens.
In this study, we report multiple novel viruses from different viral families in nasal and anal swabs collected from rodents in Hainan Province. These findings greatly increase our knowledge of the rodent-borne virus community in Hainan Province and provide basic data for analyzing the evolutionary independence of viruses. The pathogenicity and associated impact of these novel viruses on humans and animals should be evaluated in further studies.
When it comes to identifying zoonotic disease threats in the wild, we are barely scratching the surface of what is out there.
Two summers ago, in PNAS Research: Intensity and Frequency of Extreme Novel Epidemics, researchers suggested that the probability of novel disease outbreaks will likely grow three-fold in the next few decades.
Given the rate at which new zoonotic threats continue to be discovered, its hard not to consider that an optimistic assessment.