Zoonotic diseases - those which originated in or are normally hosted by non-human species, but can spill over into humans - have been with us for thousands of years.
- Tuberculosis probably jumped to humans when we began to domesticate goats and cattle 5000 years ago.
- Measles appears to have evolved from canine distemper and/or the Rinderpest virus of cattle.
- And Influenza, which is native to aquatic birds, became much more of a threat after humans domesticated ducks, geese, and chickens.
These emerging infectious diseases are considered such an important threat that the CDC maintains as special division – NCEZID (National Center for Emerging and Zoonotic Infectious Diseases) – to deal with them.
In 2014, in Emerging zoonotic viral diseases L.-F. Wang (1, 2) * & G. Crameri wrote:
The last 30 years have seen a rise in emerging infectious diseases in humans and of these over 70% are zoonotic (2, 3). Zoonotic infections are not new. They have always featured among the wide range of human diseases and most, e.g. anthrax, tuberculosis, plague, yellow fever and influenza, have come from domestic animals, poultry and livestock. However, with changes in the environment, human behaviour and habitat, increasingly these infections are emerging from wildlifeWhile coronaviruses have now joined the ranks of pandemic-producing viruses, influenza - due to its potential to reassort in numerous hosts (see graphic at the top of this page) - continues to lead the list of pandemic contenders.
Although all eyes are currently focused on H5N1 in cattle, we could easily be blindsided by any of dozens of other novel (avian, swine, canine, equine, even bat-origin) flu viruses circulating around the world.
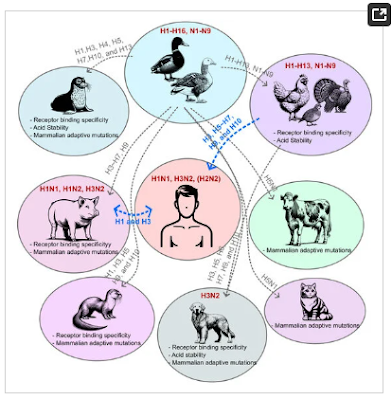
Figure 1. Cross-species transmission of influenza A virus. From their natural reservoirs in wild waterfowl, influenza A viruses (IAVs) have been transmitted to various animal species. The majority of these cross-species transmission events result in dead-end interactions between the novel IAVs and the animals (grey dashed lines, H5N1 in dairy cattle requires further studies), but some subtypes successfully circulate within that animal population (bolded red). These cross-species transmissions can also occur between animals and humans, raising public health risks (blue dashed lines). The host-adapted changes in animal-isolated IAVs are summarized below the host species.
Due to its length, I've only posted some excerpts from this review. Follow the link to read it in its entirety. I'll have a brief postscript after the break.
Abstract
The influenza A virus (IAV) has been a major cause of several pandemics, underscoring the importance of elucidating its transmission dynamics. This review investigates potential intermediate hosts in the cross-species transmission of IAV to humans, focusing on the factors that facilitate zoonotic events. We evaluate the roles of various animal hosts, including pigs, galliformes, companion animals, minks, marine mammals, and other animals, in the spread of IAV to humans.
1. Introduction
Influenza A virus (IAV) is a significant pathogen that primarily causes respiratory illnesses in humans [1]. Since its identification, IAV has been recognized as a major pathogen responsible for several notable pandemics in the 20th and 21st centuries, including the 1918 Spanish flu, the 1957 Asian flu, the 1968 Hong Kong flu, and the 2009 H1N1 pandemic [1,2]. These events underscore the virus’s capacity to cause widespread illness and significant mortality, emphasizing its critical impact on global public health.
Wild waterfowl are considered natural reservoirs of IAV, as the most diverse subtypes of the virus have been detected in these birds [3]. The segmented genome of IAV allows for frequent genetic exchange between different virus strains, leading to substantial viral diversity. This genetic reassortment, particularly during the co-infection of different IAV subtypes in the same host, accelerates the emergence of novel IAVs capable of infecting new hosts [4]. For example, the 2009 H1N1 pandemic virus (pdm09) emerged through reassortment events between human, swine, and avian IAVs [2,4]. Pigs played a crucial role in the genesis of pdm09 and its transmission to humans, underscoring the significant role of intermediate hosts in the emergence of new pandemics.
Potential intermediate hosts, such as pigs, poultry, and other animals, may serve as bridge hosts in the cross-species transmission of IAVs [4,5,6,7,8]. Similar to the case of SARS-CoV-2, where unknown intermediate hosts were vital for the virus’s adaptation and spread in humans [9], IAVs also rely on intermediate hosts to bridge transmission from their natural reservoirs to humans [6,7,8]. This review explores potential intermediate hosts of IAVs and evaluates their roles in cross-species transmission, focusing on the factors that facilitate zoonotic events.
(SNIP)
3.6. Other Mammalian Species
Other mammalian species are susceptible to avian IAVs. There have been several spillovers of avian IAVs into zoo animals, including tigers, leopards, and lions [6]. The clade 2.3.4.4b H5N1 viruses have spread along migratory flyways, resulting in genetic reassortment with local low pathogenic avian influenza viruses (LPAIs), leading to an unusually high propensity to infect mammals [150,151]. Notably, independent spillovers of the clade 2.3.4.4b H5N1 virus into red foxes occurred in 2021 and 2022 [152]. The virus isolated from the infected red foxes acquired the E627K mutation in PB2, but no further transmission between foxes was observed.
In March 2024, milk and tissue samples from dairy cattle tested positive for IAV, which was further characterized as the clade 2.3.4.4b H5N1 virus [128]. The virus infection caused mastitis in the infected cows, and evidence of cow-to-cow transmission was found [128]. In addition, domestic cats that consumed raw colostrum and milk from sick cows exhibited a fatal systemic influenza infection [128]. The viruses isolated from the infected cat were phylogenetically related to those isolated from the cows. More importantly, cow-to-human transmissions were confirmed by monitoring people exposed to the infected cows or contaminated materials including cow’s milk [153,154,155].Phylogenetic analysis revealed that this spillover was the result of a single interspecies transmission event, preceded by genetic reassortment between the clade 2.3.4.4b H5N1 and other LPAIs in wild bird species [150]. This robust genetic reassortment within highly permissive hosts increases genetic diversity and potentially contributes to interspecies transmission [156]. Some mammalian adaptive mutations, including E627K or D701N in PB2, were detected at low frequencies in the bovine H5N1 IAVs [150]. Both α-2,6-SA and α-2,3-SA glycans are expressed in the mammary gland, respiratory tract, and cerebrum of dairy cows, which might explain their susceptibility to IAVs [157]. This suggests a novel route of cross-species transmission of IAVs through the novel animal hosts, dairy cows.
4. Conclusions
Cross-species transmission of viruses from wild or domesticated animals is one of the main sources of emerging infectious agents in humans. Frequent outcomes of cross-species transmission are asymptomatic infection or dead-end events where the virus fails to establish in a new host species. However, rare but critical viral evolution in a new host can cause significant adaptations, leading to widespread outbreaks. From their natural reservoirs in wild waterfowl, IAVs have been transmitted to terrestrial birds and mammals, including humans (Figure 1). Viral adaptation in new hosts or genetic reassortment between human- and animal-origin IAVs in “mixing vessels” can lead to unpredictable changes, increasing zoonotic potential.
If it were easy for nature to generate a successful pandemic virus, we'd be hip-deep in them all of the time. Luckily, most novel viruses are evolutionary failures, unable to compete against more `biologically fit' viruses.
But it is a numbers game; the more diverse these viruses become - and the more hosts they inhabit - the better the chances are that a viable pandemic virus will emerge.
That may not happen today, or even this year. But `viral chatter' (a term popularized by David Quammen) - referring to reports of novel flu spillovers into new species - continues to grow at an unprecedented rate.
Making it more a matter of when than if.