#18,729
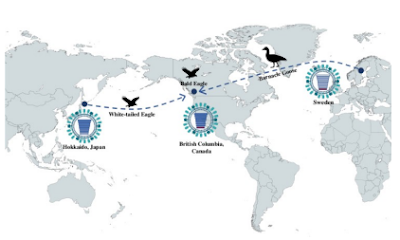
In late 2021 HPAI HNN1 arrived in Eastern Canada and (a few months later) in Western Canada via two different routes; crossing both the Pacific and the Atlantic oceans multiple times (see Multiple Introductions of H5 HPAI Viruses into Canada Via both East Asia-Australasia/Pacific & Atlantic Flyways).
Changes in the HPAI H5 clade 2.3.4.4b virus since 2016 are believed to have increased its host range (both avian and non-avian), and its ability to be carried long distances by some migratory birds (see DEFRA: The Unprecedented `Order Shift' In Wild Bird H5N1 Positives In Europe & The UK).
Within months of its arrival to North America, HPAI H5 had encountered and reassorted with numerous North American LPAI viruses, producing dozens of new genotypes of varying pathogenicity (see Preprint: Rapid Evolution of A(H5N1) Influenza Viruses After Intercontinental Spread to North America).
Here, we show that the western movement of clade 2.3.4.4b was quickly followed by reassortment with local circulating viruses, resulting in the acquisition of novel polymerase gene cassettes.
These reassortant A(H5N1) viruses are genotypically and phenotypically diverse, with many causing severe disease with dramatic neurologic involvement, in mammals.
- B3.13 aka the `bovine' strain affecting dairy cattle in at least 18 states and mildly infecting dozens of humans
- D1.1 a wild bird/poultry strain which has spilled over into > a dozen people in Washington State, severely infected a teenager in British Columbia, and produced a fatal infection in Louisiana.
- D1.2 a wild bird/poultry strain which recently detected in poultry and 2 pigs in Oregon
- D1.3 a recently detected wild bird/poultry strain which has been infection poultry and at least 1 human
This week the EID Journal published a Dispatch characterizing this recently arrived D1.1 genotype, and discussing ways it could have evolved. As you'll see, there are still a number of unknowns.
Highly Pathogenic Avian Influenza A(H5N1) in Wild Birds and a Human, British Columbia, Canada, 2024
Chelsea G. Himsworth, Jessica M. Caleta, Agatha N. Jassem, Kevin C. Yang, James E.A. Zlosnik, John R. Tyson, Laurie Wilson, Kevin S. Kuchinski, Jolene Giacinti, Megan Willie, Tony D. Redford, Maeve Winchester, Caeley Thacker, Yohannes Berhane, Theresa Burns, Natalie Prystajecky, and Shannon L. Russell
Abstract
We characterized highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b genotype D1.1 in wild birds and a human in British Columbia, Canada, during 2024. D1.1, the predominant genotype circulating in fall 2024, is a reassortment between Eurasian A3 lineage viruses, introduced to North America in 2022, and North American lineage viruses.
In fall 2021, highly pathogenic avian influenza (HPAI) A(H5N1) virus clade 2.3.4.4b was introduced into wild birds and domestic poultry in eastern Canada via the East Atlantic Flyway (1). It subsequently spread throughout North America before arriving in British Columbia, Canada, in April 2022 (1). A second incursion of HPAI H5N1 virus, clade 2.3.4.4b, brought in by the Pacific Flyway (genotype A3) (2), occurred in February 2022, resulting in both viruses circulating among wild birds in the province and causing numerous spillover events into poultry (3). The virus affected more poultry flocks in British Columbia than in any other province in Canada (4), likely because high-density poultry farming is co-located with optimal habitat for overwintering waterfowl in the Fraser Valley (3).
As of the end of 2024, British Columbia had endured 4 distinct wave of HPAI H5N1 clade 2.3.4.4b virus; each wave was characterized by increased detections in wildlife and poultry, the emergence of new genotypes (Table), and shifts in the dominant genotype (Figure 1). Most circulating HPAI H5N1 clade 2.3.4.4b viruses in the first 3 waves were reassortant descendants of the virus introduced via the East Atlantic Flyway (3); however, in fall 2024, a new wave of infections occurred in British Columbia wild bird populations associated primarily with a novel genotype (D1.1, 3) that was a descendant of the A3 genotype. We used whole-genome sequencing and phylogenetic analysis of HPAI H5N1 clade 2.3.4.4b viruses detected in wild birds in British Columbia during October and November 2024 to describe the features, ecology, and possible origins of this genotype.
(SNIP)
Conclusions
The once dominant B genotypes associated with the HPAI H5N1 clade 2.3.4.4b virus incursion via the Eastern Atlantic Flyway do not appear to be circulating in wild birds within the Pacific Flyway as of fall 2024 (despite the continuing presence of genotype B3.13 in cattle in the western United States [8]) (Table).
Instead, a novel genotype, D1.1, has emerged that is the result of a reassortment among the A3 genotypes originally introduced via the Pacific Flyway and >1 North American lineage avian influenza viruses. This virus spilled over into poultry in British Columbia and infected 1 human and has also been detected in parts of the United States south of British Columbia (Figure 2).
Compared with A3 viruses detected a year earlier, the hemagglutinin segment of the D1.1 viruses had acquired fewer net substitutions than expected, despite sharing a relatively recent common ancestor (Figure 1, panel B). This finding could suggest that the D1.1 genotype or its ancestors may have been preserved in an environmental reservoir—e.g., in frozen wetlands in the high arctic (9)—in the summer of 2024 before being reintroduced into migratory birds in the fall.
Alternatively, this viral lineage may be particularly well adapted to certain wild bird species or populations, resulting in circulation with minimal evolutionary pressure. This finding has implications for the use of molecular clock theory in phylodynamic modeling of HPAI viruses.
We note that D1.1 appears to be unique among the HPAI H5N1 clade 2.3.4.4b genotypes due to the acquisition of a North American lineage NA segment. The Am4N1 NA segment likely originated from a reassortment event involving waterfowl in western Canada, potentially within British Columbia. Further studies are needed to determine where and when the other reassorted segments were acquired.
Of interest, the prevalence of environmental HPAI H5N1 virus clade 2.3.4.4b detections based on genomic analysis of wetland sediment was far greater in fall 2024 than for data from fall 2023 (10). This phenomenon could suggest that a great number of birds were infected with D1.1 compared with other genotypes in previous years, that the genotype is associated with greater viral shedding, or both.
The NA segment encodes the enzyme required for viral release from infected cells (11). Certain NA lineages, therefore, might increase viral shedding in wild birds. Considering the explosive wave of poultry outbreaks observed in British Columbia in late 2024, it would be prudent to investigate whether the Am4N1 NA segment has a functional impact on host-virus interactions. In addition, it will be important to determine the implications of D1.1 for host range and infectivity, given that the virus detected in the human case was most closely related to those found in wild birds, suggesting the potential for direct or indirect transmission from wild birds to humans.
Dr. Himsworth is a veterinary pathologist and epidemiologist who is the Deputy Chief Veterinarian for the Province of British Columbia, Canada, the British Columbia Regional Director for the Canadian Wildlife Health Cooperative, and an Associate Clinical Professor in the School of Population and Public Health at the University of British Columbia. Her research and practice centers around One Health–based transdisciplinary approaches to the surveillance and management of health issues associated with wild and domestic animals.
The emergence of B3.13 in cattle in early 2024, followed by the arrival of an aggressive D1.1 last fall, illustrate just how quickly the avian `fluscape' can change.
Add in the fact that globally there are numerous HPAI H5 variants circulating in different hosts (e.g. marine mammals, livestock, peridomestic animals, wild birds and poultry) - each on their own evolutionary path - and you have a lot of potential contenders.
A 2016 study (see Sci Repts.: Southward Autumn Migration Of Waterfowl Facilitates Transmission Of HPAI H5N1), suggests that waterfowl pick up new HPAI viruses in the spring (likely from poultry or terrestrial birds) on their northbound trip to their summer breeding spots - where they spread and potentially evolve - and then redistribute them on their southbound journey the following fall.Migratory birds are now headed back to their high latitude summer breeding areas in Alaska, Siberia, and above the Arctic circle. But they will begin their southbound migration in a few short months.
Between antigenic drift and antigenic shift (reassortment), the HPAI H5Nx viruses that return next fall could easily be more - or less - of threat than what we've seen thus far. Evolution is a double-edged sword, and what it adds it can also take away.
But one thing is certain; evolution never stops. Which is why - even if we should enjoy a relatively quiet avian flu summer - we should be prepared for another round of surprises in the fall.