# 5531
When the 2009 H1N1 virus emerged just over two years ago, the assumption was that it would begin to evolve (or mutate) fairly rapidly.
There were real concerns that it might quickly pick up oseltamivir (Tamiflu) resistance, and even some worries it might hook up (reassort) with the H5N1 avian flu and produce some kind of Frankenswine Virus.
Instead, for the first 12 months or so, we received assurances from the CDC and the World Health Organization (WHO) that the virus was unusually stable.
What few variations that were seen were reportedly antigenically very similar to the A/California/7/2009 H1N1 virus that the vaccine was based upon.
But of course, some changes, and variations, were observed.
There were a handful of oseltamivir resistant cases reported – most (but not all) appeared to have developed spontaneously in a patient actually taking the drug.
During a 14-month period (April 2009-June 2010) 6,740 H1N1 samples were submitted to US surveillance systems for testing, and of those, only 37 (.5%) proved resistant to oseltamivir.
This kind of resistance is usually caused by a mutation (H275Y) where a single amino acid substitution (histidine (H) to tyrosine (Y)) occurs at neuraminidase position 275.
* * * * * *
The D225G `Norway’ mutation made headlines in November of 2009, but it was observed in both mild and severe cases, and so it wasn’t at all clear what the clinical significance was (see Eurosurveillance: Debating The D222G/N Mutation In H1N1).
But with millions of infected hosts (people/birds/pigs) replicating trillions of copies of the virus every day, mutations were inevitable, and viral evolution was bound to take hold.
Many were `flashes in the pan’, and due to inferior biological fitness, failed to propagate well. But by the middle of 2010, we began to see some subclades of the 2009 H1N1 virus that had exhibited some traction.
Notably the A/Hong Kong/2213/2010 and the A/Christchurch/16/2010 (highlighted by D222N) subgroups.
In September of 2010, the WHO Influenza Centre in London released an analysis of the evolution of the H1N1 and seasonal viruses to be used in deciding the makeup of the Southern Hemisphere 2011 flu vaccine.
They acknowledged these new subgroups (and others), but stated (bolding mine):
The A(H1N1) pandemic 2009 viruses propagated at NIMR remain antigenically similar to the vaccine virus A/California/7/2009. Fewer low reactors have been detected in 2010 than was observed in 2009.
New genetic sub-clades have been detected but they do not appear antigenically distinct from the majority of A(H1N1) pandemic 2009 viruses collected since the start of the pandemic.
Not quite the same thing as saying that all of the viruses collected are antigenically close to the vaccine strain. But we live in an imperfect world.
Herein lies the dilemma for those who must choose which virus strains to include in a vaccine 6 months before it can be deployed.
The influenza virus is a constantly moving target.
And unlike a school of fish, that all change direction at at the same time, flu viruses go their own way. It’s very messy. And very difficult to predict.
Worse, it is entirely possible that you can have a field of viruses circulating with enough antigenic diversity that not all of them can be covered by the vaccine. The best you can hope for is to include the most prevalent strains.
Hence the occasional reports of `vaccine escapes’, and `low reactors’.
Actually we see a similar situation every year when the vaccine committees choose one of the two B viruses (Yamagata or Victoria strain) to include in the vaccine. Some people who take the flu vaccine will be unlucky enough to catch the strain not included that year.
Flu vaccines, most years, are pretty good. But they aren’t perfect. And some years they miss the mark badly.
As I tell people, if you want a guarantee . . . buy a Craftsman.
By late 2010 another subgroup A/England/142/2010 began to spread widely across Europe.
By the end of the 2010-2011 flu season (week 16), at least in the European theatre – its prevalence had nearly equaled the number of A/California/7/2009-like isolates detected.

Chart based on data from Euroflu Report Week 16 : 18/04/2011-24/04/2011
In other parts of the world, the A/Hong Kong and A/Christchurch strains had much larger shares of the influenza pie.
As far as antiviral resistance goes (again from the WHO surveillance of Europe) we find that it is only slowly increasing, with the overwhelming majority of isolates tested still showing sensitivity to Tamiflu.
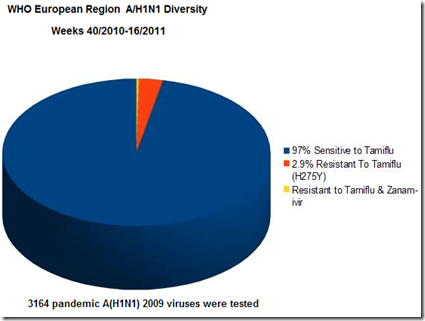
But as we move further downrange from the emergence of the 2009 H1N1 virus, the odds are that we’ll see more diversity in the subgroups, more `low reactors’, and more antiviral resistance.
If a virus – which leaves behind immunity in the host – fails to evolve into a new antigenically different strain, it will eventually die out due to a lack of susceptible hosts.
Survival demands that the virus evolve.
And in recent months we’ve seen reports – particularly out of Europe – suggesting that may be happening, as the effectiveness of the seasonal flu vaccine over the 2010-2011 flu season had dropped.
CIDRAP covered this report in late March.
Preliminary studies show lower flu-shot effectiveness in Europe
Robert Roos
News Editor
Mar 21, 2011 (CIDRAP News) – Preliminary studies suggest that this year's trivalent seasonal flu vaccine used in Europe was less effective against the 2009 H1N1 virus than last year's monovalent H1N1 vaccine was, possibly because of some degree of mutation in the virus, according to recent reports in Eurosurveillance.
Which brings us to an interesting report, mentioned by CIDRAP last night in their news roundup, coming from the US Department of Defense (DoD).
It states that recent outbreaks of H1N1 in Venezuela and Mexico's Chihuahua state need to be monitored because they has been linked to severe infections and deaths, including some patients who had received the vaccine.
Initial analysis are suggesting that the Mexico group (inDRE1945) of H1N1 does not fall into previously characterized A/England/142/2010, A/Christchurch/16/2010 (highlighted by D222N), or A/Hong Kong/2213/2010 subclades.
http://airforcemedicine.afms.mil/idc/groups/public/documents/afms/ctb_152827.pdf
These outbreaks have been well monitored for more than a month by the newshounds on the flu forums, including in this thread on FluTrackers, which has more than 250 entries.
But what to make of all this is less than clear.
Sub clades of circulating influenza viruses emerge all the time. Some disappear almost immediately, others linger in the background for awhile, sputter and die.
A very few take off like a rocket.
It is a case of survival of the fittest. The virus that evades acquired immunity the best, replicates well, and transmits the most efficiently usually wins the race.
At least for a while.
In the world of influenza viruses, nothing is permanent, the status quo never lasts for long, and the only constant is change.
Obviously, anything that increases the virulence, or moves the virus away from our acquired immunity (through prior infection or vaccines), is of concern.
But whether this suspected branch in H1N1’s evolution in Mexico & Latin America proves to have `legs’, remains to be seen.
It is possible that after a relatively mild second year of A/H1N1/2009, we could see a more severe flu season come the fall due to evolution of the virus.
It’s happened before.
In 1957, the Asian Flu pandemic seemed to disappear completely for more than a year, only to return in 1959 and again after a two year lull during the 1962-63 flu season.
With the Northern Hemisphere’s flu season at an end, we’ll be looking to the events south of the equator over the next six months to give us some hint of what may be in store for next fall.
Stay tuned.

