Recombination is the sharing of genetic material between similar genomes that simultaneously infect a single host. The resultant hybrid is called a recombinant.
Recombination events have undoubtedly occurred with COVID since the beginning, but they have become easier to identify since the emergence of Omicron, which differs enough genetically from earlier COVID variants (Alpha, Delta, `Wild type') to make Delta/Omicron hybrids stand out.
But as early as May of 2020 we were seeing signs of recombination (see More COVID-19 (SARS-CoV-2) Mutation Reports)); signs which grew stronger in 2021 with the emergence of both Alpha and Beta variants (see ECCMID: Dual Infection With COVID Variants (Alpha & Beta)).
Also in the spring of 2021, a preprint published by researchers from the Viral Diseases Branch, Walter Reed Army Institute of Research, MD, USA, found moderate evidence for 8 SARS-CoV-2 recombination events (see A comparative recombination analysis of human coronaviruses and implications for the SARS-CoV-2 pandemic).
In July of 2021, in UK SAGE: Can We Predict the Limits of SARS-CoV-2 Variants and their Phenotypic Consequences?, UK scientists discussed at length the unpredictable ramifications that new recombinant COVID variants could have on the future course of the pandemic.
That said, most variants are evolutionary failures.
They emerge, and may even flourish briefly, but eventually are overpowered by more biologically `fit' viruses. There have been literally thousands of failed COVID variants for every `Delta' or `Omicron' success story. And yes, some of these have been recombinants.
While several recombinant COVID variants are now being tracked by the WHO, ECDC and the UKHSA, it will take some time to determine if any of these can compete with existing strains. And if they can, whether they pose any greater (or lesser) risk to public health.
Earlier this week the WHO announced that they were now monitoring 3 recombinant COVID variants (XD, XE & XF), although they cautioned:
None of the preliminary available evidence indicates that these recombinant variants are associated with higher transmissibility or more severe outcomes.
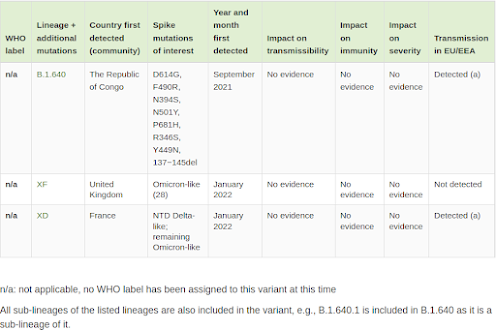
Variants under monitoring
These additional variants of SARS-CoV-2 have been detected as signals through epidemic intelligence, rules-based genomic variant screening, or preliminary scientific evidence. There is some indication that they could have properties similar to those of a VOC, but the evidence is weak or has not yet been assessed by ECDC. Variants listed here must be present in at least one outbreak, detected in a community within the EU/EEA, or there must be evidence that there is community transmission of the variant elsewhere in the world.
While data on these recombinant variants is very limited, the following UKHSA COVID Technical report provides us with perhaps the most detailed information to date. Follow the link for all of the charts and graphs, and an update on Omicron BA.2.
25 March 2022
(Excerpts)
Signals from horizon scanning: Recombinant lineages XD, XE, XF
Three recombinant lineages have been designated by Pangolin. Two are a combination of Delta and BA.1 (XD and XF). XD, which has an Omicron S gene incorporated into a Delta genome, requires biological characterisation. It is present in several European countries but has not been detected in the UK. XF caused a small cluster in the UK but has not been detected since 15 February. XE is a BA.1/BA.2 recombinant, with the majority of the genome including the S gene belonging to BA.2. XE shows evidence of community transmission within England, although it is currently <1% of total sequenced cases. Early growth rates for XE were not significantly different from BA.2, but using the most recent data up to 16 March 2022, XE has a growth rate 9.8% above that of BA.2. As this estimate has not remained consistent as new data have been added, it cannot yet be interpreted as an estimate of growth advantage for the recombinant. Numbers were too small for the XE recombinant to be analysed by region.
(SNIP)
Part 2. Signals from horizon scanning 2.1
Newly designated recombinantsThere are currently 3 recombinant lineages being monitored as part of horizon scanning: XD, XE, and XF (Figure 6). XD and XF are Delta and BA.1 recombinants. XE is a BA.1 and BA.2 recombinant and has 3 mutations that are not present in all BA.1 or BA.2 sequences: NSP3 C3241T and V1069I, and NSP12 C14599T. XF and XE are associated with UK sequenced samples.
XD is predominantly associated with France. XD contains the unique mutation NSP2: E172D. Figure 6 shows the SARS-CoV-2 genomes full length (first tier, orange), gene locations across the genome (second tier, dark orange, yellow, and orange). Reference genomes colours: fourth tier Delta (Green), fifth tier BA.1 (purple), and sixth tier BA.2 (lilac). XD, XE and XF are in tiers 7 to 9 are coloured by the associated references, annotated with break points and the gene location of their break points. On the right hand side, the GitHub Pangolin lineage request number is listed. Figure 6. XD, XE and XF – newly designated recombinant lineages

(SNIP)
2.2 Epidemiology of XDThe XD recombinant lineage is a Delta AY.4 genome that has acquired a BA.1 spike sequence (nucleotide positions 21,643 to 25,581). As of 22 March 2022, XD has not been detected in the UK. Screening GISAID indicates that there are 49 samples that meet the XD definition and show no signs of contamination. The earliest collection date for these samples is the 13 December 2021 and the most recent collection date is 13 March 2022 (Figure 7). Forty samples have been identified from France, 8 from Denmark, and 1 from Belgium (Table 1).
2.3 Epidemiology of XFThe XF lineage is a recombinant of Delta and BA.1 with a break point near the end of NSP3 (nucleotide 5,386). In total 39 UK sequenced samples were identified and validated as part of the XF recombinant lineage. The earliest known sample date in the UK dataset is 7 January 2022 and the most recent sample to date is 14 February 2022 with no recent evidence for growth (Figure 8). Given the lack of evidence for recent UK samples from this lineage it is unlikely to be associated with sustained community growth. There is currently no evidence for samples from non-UK countries on GISAID for this lineage.
2.4 Epidemiology of XE
The XE recombinant contains BA.1 mutations for NSP1-6 and then BA.2 mutations for the remainder of the genome. As of 22 March 2022, there are 763 XE sequences in the UK data. As of 22 March 2022, there are 637 XE cases in England. These are geographically distributed across England and increasing in number, with the first case detected via sequencing on 19 January 2022, and most cases in East of England, London, and the South East (Figure 11).
Although it is very early days, XE appears to be the most transmissible of the three. But it will take weeks, perhaps months, before we know whether any of these variants will pose a significant public health threat, or if they will all end up as obscure footnotes to this pandemic.
Stay tuned.