#16,719
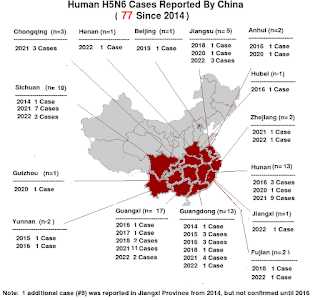
China's avian H5N6 virus, which has been on our radar since the spring of 2014 when it infected a man in Sichuan Province, caused just over 2 dozen known human infections during its first six years of circulation (average of 4 per year).
As with all influenza subtypes, when we talk about H5N6, we're aren't talking about a single viral threat, but rather an expanding array of related viruses sharing similar HA and NA genes. Viruses which can vary considerably in their behavior and the threat they pose (see Differences In Virulence Between Closely Related H5N1 Strains).Since the fall of 2020, however, we've seen a marked increase in cases, with more than 50 reported over the past 18 months (see map above). This number, however, is likely an undercount.
Although categorized by their two surface proteins (HA & NA) Influenza A viruses have 8 gene segments (PB2, PB1, PA, HA, NP, NA, M1, M2, NS1, NS2).

Shift, or reassortment, happens when two different influenza viruses co-infect the same host swap genetic material. New hybrid viruses may be the result of multiple reassortments, with gene contributions coming from several parental viruses.
China CCDC Weekly: Genetic Characterization of Two Human A (H5N6) Viruses — Guangxi , China, 2021
CCDC Weekly: Outbreak Report - Five Independent Cases of Human Infection With HPAI H5N6 — Sichuan Province
For now, H5N6 has shown no ability to spread efficiently from human-to-human. And as long as that status quo remains, the virus is primarily a threat to those who have close contact with poultry in China.
The following research article, published yesterday in the EID Journal, reports on 13 types of reassortant H5N6 viruses that have infected humans in China - 4 of them added since 2021 - and on the detection of several mammalian-adapted mutations.
Research
Abstract
The recent rise in the frequency of influenza A(H5N6) infections in China has raised serious concerns about whether the risk for human infection has increased. We surveyed epidemiologic, clinical, and genetic data of human infections with A(H5N6) viruses. Severe disease occurred in 93.8% of cases, and the fatality rate was 55.4%. Median patient age was 51 years.
Most H5N6 hemagglutinin (HA) genes in human isolates in 2021 originated from subclade 2.3.4.4b; we estimated the time to most recent common ancestor as June 16, 2020. A total of 13 genotypes with HA genes from multiple subclades in clade 2.3.4.4 were identified in human isolates. Of note, 4 new genotypes detected in 2021 were the major causes of increased H5N6 virus infections. Mammalian-adapted mutations were found in HA and internal genes.
Although we found no evidence of human-to-human transmission, continuous evolution of H5N6 viruses may increase the risk for human infections.
(SNIP)
Discussion
Among AIVs, viruses of subtypes H5 and H7 have been of particular concern because of their high rates of death. The hospitalization fatality risk for H5N6 infections (59.0%) was slightly lower than that for H5N1 infections (70.0%) but higher than that for H7N9 (35.0%) (44). In addition, the median age of patients with H5N6 virus (51 years) was older than the median age for patients with H5N1 virus (26 years) but younger than the age for patients with H7N9 virus (62 years) (44).
On the basis of epidemiologic investigations, 93.8% of influenza H5N6 case-patients were confirmed to have poultry exposure history. The contamination of live poultry markets and backyard birds, as well as the practice of processing poultry without personal protection, could be ongoing exposure sources for influenza A(H5N6) virus. While H5N6 viruses continue to circulate in poultry, human infections will undoubtedly continue.
WHO has recommended early antiviral therapy, ideally within 48 hours of symptom onset, for suspected or confirmed influenza patients (45). Our study found that early initiation of antiviral treatments could reduce the fatality rate in H5N6 patients to some extent. However, symptoms at the onset of influenza H5N6 infections were clinically similar to those of other respiratory pathogen infections. Thus, increased sensitivity of diagnostic systems is needed to improve case identification and initiate timely antiviral treatment.
During the COVID-19 pandemic, diagnostic capacity for respiratory illnesses among human health systems in China, including hospitals, Centers for Disease Control and Prevention at different levels, and third-party agencies, was increased. In our study, clinical samples from 15 H5N6 cases during the COVID-19 pandemic (2 in 2020 and 13 in 2021) were first identified by third party agencies. This additional diagnostic capacity contributed to the detection of H5N6 cases reported in 2021.
Genetic analysis in our study detected at least 13 types of reassortant H5N6 viruses in infected humans in China (Figure 6). Origins of internal genes were dramatically diversified, indicating the advanced genetic compatibility of H5N6 viruses with other AIVs. In 2021, a total of 4 new H5N6 genotype viruses emerged and accounted for almost all of the H5N6 human viruses based on the available full genome. Of note, different from previously isolated viruses, the HA gene of almost all H5N6 human viruses in 2021 belonged to genetic clade 2.3.4.4b and was closely related to that of the first H5N8 isolate, which caused infection in a patient in Russia (46). Additional mammal-adapted mutations, including Q226L and T192I in the HA protein, which could increase the viral affinity for human cells, were also detected (Appendix Table 3), indicating the viral adaptation process from birds to humans.
In summary, although we observed a rise in the number of influenza A(H5N6) infections in 2021, the disease course and CFR were comparable to previously detected H5N6 cases. Antiviral drugs remain effective if used early. However, new genotype viruses and mammal-adapted substitutions emerged. Moreover, reports from OFFLU (https://www.offlu.org ) and WHO have documented that clade 2.3.4.4h and 2.3.4.4b H5N6 viruses and clade 2.3.4.4b H5N8 viruses have been detected in poultry and wild birds in China.
Considering the continuous viral circulation in birds and incidence of human infection, more H5N6 variants and genotypes with further advantages in humans might emerge. Increased attention to such emerging viruses is vital for public health and pandemic preparedness.
Dr. Zhu works in the Chinese National Influenza Center, National Institute for Viral Disease Control and Prevention, China Center for Disease Control and Prevention. Her research interests include evolutionary analysis and pathogenicity mechanism of influenza viruses.
Of course, until quite recently, we didn't have proof that a novel coronavirus could spark a human pandemic either.
It is probably more likely that the next influenza pandemic will emerge from an avian or swine H1, H3, or H3 virus that carries a much lower case fatality rate. The CDC currently ranks a Chinese Swine-variant EA H1N1 `G4' as having the highest pandemic potential of any flu virus on their list.
But another pandemic is inevitable, and like it or not, long-shots sometimes do come in.