#17,453
A week ago, in Netherlands: Zoonoses Experts Council (DB-Z) Risk Assessment & Warning of Swine As `Mixing Vessels' For Avian Flu, we looked at growing concerns that avian H5N1 - which has shown a growing predilection for spilling over into mammalian hosts - could increase its pandemic threat by spreading (and evolving) in farmed swine.
This was also a concern voiced 2 months ago, when the ECDC/EFSA Avian Influenza Overview December 2022 – March 2023 warned:
The additional reports of transmission events to and potentially between mammals, e.g. mink, sea lion, seals, foxes and other carnivores as well as seroepidemiological evidence of transmission to wild boar and domestic pigs, associated with evolutionary processes including mammalian adaptation are of concern and need to be closely followed up.
While field detections have been limited, over the years we've seen evidence that H5N1 can infect pigs, albeit usually asymptomatically. A few past reports include:
WHO H5N1 detected in pigs in China (2004)
EID Journal: Asymptomatic H5N1 In Pigs (2010)
An Unusual Report Of H5N1 in Pigs (Indonesia 2016)
Sci. Rpts.: Evidence Of H5N1 Exposure In Domestic Pigs - Nigeria (2018)
Between extremely limited testing, and the fact that H5N1 tends to be asymptomatic (or mildly symptomatic) in pigs, it is probably more common than we realize.
All of which brings us to a recent report published the journal Microorganisms, which describes a spillover event at a `mixed species' farm (poultry & swine) in Italy, and the subsequent seroconversion of the majority of the pigs tested on that farm.
While the authors speculate that a common exposure, rather than swine-to-swine transmission, led to the large number of seropositive pigs, this still presents the virus with new opportunities to better adapt to a mammalian host, or to reassort with other viruses.
Due to its length I just posted the link, and a few excerpts. The full study is well worth reading. I'll have a brief postscript when you return.
Francesca Rosone 1,*,†,Francesco Bonfante 2,†, Marcello Giovanni Sala 1,†, Silvia Maniero 2, Antonella Cersini 1, Ida Ricci 1,Luisa Garofalo 1,Daniela Caciolo 1, Antonella Denisi 1, Alessandra Napolitan 2, Monja Parente 3, Bianca Zecchin 2, Calogero Terregino 2,* and Maria Teresa Scicluna 1
Microorganisms 2023, 11(5), 1162; https://doi.org/10.3390/microorganisms11051162 Published: 28 April 2023
Abstract
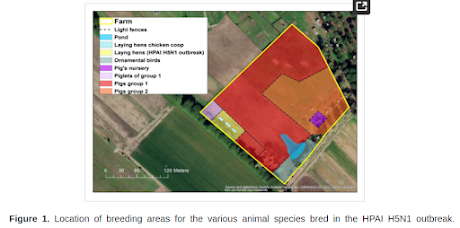
Starting from October 2021, several outbreaks of highly pathogenic avian influenza virus (HPAIV) subtype H5N1 were reported in wild and domestic birds in Italy. Following the detection of an HPAIV in a free-ranging poultry farm in Ostia, province of Rome, despite the lack of clinical signs, additional virological and serological analyses were conducted on samples collected from free-ranging pigs, reared in the same holding, due to their direct contact with the infected poultry.
While the swine nasal swabs were all RT-PCR negative for the influenza type A matrix (M) gene, the majority (%) of the tested pigs resulted serologically positive for the hemagglutination inhibition test and microneutralization assay, using an H5N1 strain considered to be homologous to the virus detected in the farm.
These results provide further evidence of the worrisome replicative fitness that HPAI H5Nx viruses of the 2.3.4.4b clade have in mammalian species. Moreover, our report calls for additional active surveillance, to promptly intercept occasional spillover transmissions to domestic mammals in close contact with HPAI affected birds. Strengthened biosecurity measures and efficient separation should be prioritized in mixed-species farms in areas at risk of HPAI introduction.
(SNIP)
Discussion and ConclusionsIn this report, we describe a multi-species outbreak caused by a 2.3.4.4b clade H5N1 HPAI virus in a rural farm in the province of Rome, Italy. The virus was detected in laying hens severely affected by the infection accompanied by high mortality, while geese, ducks and pigs in the same farm, although separated by light fences, remained healthy throughout the investigation. We could not detect viral genome in the nasal swabs collected from any of the sampled pigs, but we observed a robust seroconversion of the herd and in the few tested geese to an H5N1 HPAI virus homologous to the strain detected in the laying hens.(SNIP)Interestingly, the observed phenotype of infection appears to be milder than the ones described in previous experimental work based on other goose/Guangdong (gs/GD) lineage viruses of clade 0, 1, 2.1, 2.2 and 2.3, which induced moderate clinical manifestation, reaching low to moderate infectious titers at the level of the nose [15,38,39]. Irrespective of the genetic clade used for the challenge, all of these studies consistently reported lack of transmission to contact animals, indicating a low adaptation of the gs/GD lineage in this species.In light of this, we speculate that the high incidence of seroconversion observed among pigs during this outbreak is unlikely to be the result of secondary spread within the herd, rather the consequence of a prolonged exposure of the animals to high viral loads through fomites, contaminated soil and consumption of contaminated feed. In light of this, we hypothesize that the seroconversion observed among pigs during this outbreak is probably the result of secondary spread within the herd, via high contaminated fomites, soil or feed from infected birds.Unfortunately, the data at our disposal were not sufficient to reach a conclusion on the intra-farm transmission chain.Similar ecological and pathogenic drivers might have underpinned the HPAI H5N1 infections and seropositivity described in commercial and rural swine in Nigeria [14] China [40] and Indonesia [41], where endemicity for HPAI viruses in poultry favored spillover events to domestic mammals.Interestingly, in one of these studies, Meseko et al. [14] found HI titers against a 2.3.2.1c clade H5N1 virus comparable to the ones reported in our study, despite recording in healthy animals only high Ct values at the level of the trachea in an abattoir.Our investigation had several limitations.
- Virological sampling in pigs only took place about a week after the serological sampling, and about 17 days after the virus had led to almost 100% mortality in the flock of laying hens. Assuming that no secondary transmission occurred among pigs, the diagnostic sensitivity of our virological investigation was probably impaired by the delayed sampling.
- Moreover, for the logistical complexity inherent to the free-range setting of the farm, only adult pigs were sampled for blood, while piglets were not evaluated. Additionally, our approach to differential serology might have suffered from suboptimal sensitivity, due to the fact that the swine influenza antigens we used were not the most recent.
- This might explain in part the negative HI results for both avian and swine influenza in 19 pigs that had anti-NP antibodies. Due to our inability to detect and sequence the virus in pigs, we could not formulate a complete risk assessment, since molecular analyses necessary to identify markers of adaptation and virulence were only based on the virus identified in poultry in the same premise.
- Nonetheless, based on the negative rRT-PCR results on nasal swabs, we consider unlikely the possibility that a mammalian well-adapted reassortant strain between a swine influenza virus and the H5N1 virus was generated. The absence of symptoms and mortality in pigs in this and other studies is in stark contrast with the frequent reports of neurological syndromes associated with infections in wild mammals in Europe and North America, where 2.3.4.4b viruses have reached unprecedented levels of circulation [42].
- This discrepancy might depend on the different anatomical route of infection, viral load, host susceptibility and the specific genotype responsible for the outbreak. Active serosurveillance in wildlife and farmed pigs will be key to understand the susceptibility of these species to infection with 2.3.4.4b H5Nx viruses.
- The reassortment of swine and H5Nx viruses of the Gs/GD lineage was previously reported [43] and poses a considerable risk for the emergence of zoonotic and prepandemic viruses, as demonstrated by the efficient aerosol transmission in guinea-pigs of hybrid HPAI H5N1 viruses carrying human-like polymerase and non-structural proteins [44].
In conclusion, we advocate for additional monitoring of swine in close proximity or exposed to HPAI infected poultry, to achieve a timely identification of spillover events, given the delicate role played by this species in the emergence of reassortant zoonotic viruses. Functional separation of poultry species and mammals should be prioritized as a preventive measure to reduce the risk of inter-species transmission.
(Continue . . . )
This event occurred fairly early (Oct 2021) in the current wave of HPAI H5N1, before its spread to the Americas, and prior to most of the spillover reports we've seen.
According to the latest ECDC/EFSA Avian flu surveillance report - more than a dozen new genotypes have emerged in Europe since then - with even more identified in Asia and in the Americas.
The virus marches on, and while the lack of evidence for swine-to-swine transmission in this report is comforting, it tells us very little about the current - or future - capabilities of HPAI H5N1.
The growing diversity of H5N1 in the wild, and the increasing frequency of mammalian infections, provides the virus with new opportunities to adapt (or reassort) into a bigger threat (see below). While none of that guarantees a bad outcome, it is hard to see either as a positive development.
, 
For more on the risks of `mixed farming' and the zoonotic spread of viruses, you may wish to revisit:
Viruses: Zoonotic Animal Influenza Virus and Potential Mixing Vessel Hosts
Eurosurveillance: HPAI A(H5N1) Virus Infection in Farmed Minks, Spain, October 2022