#17,674
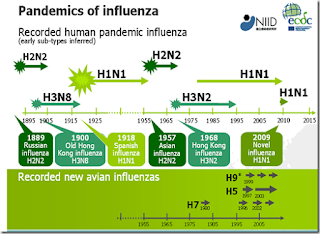
While it is always possible that an exotic avian influenza virus like H5N1 could adapt to humans, all of the known influenza pandemics over the past 130 years have come from H1, H2, and H3 viruses (see graphic above).
Admittedly, things get a bit murky when you go back past 1918, but it appears that H1, H2, and H3 influenza viruses have an easier time jumping to humans. These subtypes are common in both birds and swine, and the 2009 H1N1 pandemic was caused by a triple-reassortant swine flu virus.
Every year a small number of `swine-variant' infections are reported in humans in the United States, and around the world, but they undoubtedly occur more often than we know. Most people who develop flu-like symptoms are never tested, and so many `novel' cases likely fly under the radar.
The CDC's risk assessment for Swine Variant viruses reads:
Sporadic infections and even localized outbreaks among people with variant influenza viruses may occur. All influenza viruses have the capacity to change and it’s possible that variant viruses may change such that they infect people easily and spread easily from person-to-person. The Centers for Disease Control and Prevention (CDC) continues to monitor closely for variant influenza virus infections and will report cases of H3N2v and other variant influenza viruses weekly in FluView and on the case count tables on this website
Most swine variant infections are contracted via direct contact with pigs - by farmers, and sometimes attendees at agricultural fairs (see CDC FluView On 2 Swine Variant Infections (H3 & H1N2v)) - but sometimes there is no obvious swine exposure.
The risk of one of these swine variant viruses sparking a pandemic is relatively low, but it isn't zero. The CDC's IRAT (Influenza Risk Assessment Tool) lists 3 North American swine viruses as having at least some pandemic potential (2 added in 2019).
H1N2 variant [A/California/62/2018] Jul 2019 5.8 5.7 Moderate
H3N2 variant [A/Ohio/13/2017] Jul 2019 6.6 5.8 Moderate
H3N2 variant [A/Indiana/08/2011] Dec 2012 6.0 4.5 Moderate
Overnight the World Health Organization published a DON report on a recent Swine H1N1 infection in the Netherlands, one where there was no clear exposure to pigs. I've only posted some excerpts, so follow the link to read it in its entirety.
Influenza A (H1N1) variant virus - the Netherlands
13 September 2023
Situation at a glance
On 2 September 2023, the Ministry of Health, Welfare and Sport of the Netherlands notified the World Health Organization (WHO) of a laboratory-confirmed human case of infection with a swine-origin influenza A(H1N1) variant (v) virus in the province of North Brabant, Netherlands. This is the first human infection caused by influenza A(H1N1)v virus reported in the Netherlands in 2023.
Worldwide sporadic human cases of influenza A(H1N1)v have been reported previously, including from the Netherlands. According to the International Health Regulations (IHR,2005), a human infection caused by a novel influenza A virus subtype is an event that has the potential for high public health impact and must be notified to the WHO.
This case was picked up as part of routine surveillance of respiratory illnesses. Based on the available information, there is no clear indication of the source of infection, and no direct contact with pigs was reported. As of 7 September, there were no symptomatic contacts of this case and no further detections have been reported in routine surveillance. All five close contacts were followed for 10 days- the maximum incubation period and none developed symptoms. Thus, there was no evidence of person-to-person transmission and the case is considered as a sporadic human case of influenza A(H1N1)v. The likelihood of community-level spread among humans and/or international disease spread through humans is considered as low.
Description of the case
On 2 September 2023, WHO was notified of a confirmed human infection with a swine influenza A(H1N1)v virus, in Netherlands through the European Commission's confidential Early Warning and Response System (EWRS).
The case is an adult from the province of North Brabant with no underlying medical conditions and no history of occupational exposure to animals. On 20 August 2023, the patient developed fatigue and general malaise and the following day developed an acute respiratory infection with onset of chills, sneeze, cough, headache, and generalized weakness, followed by fever on 22 August.
On 21 August 2023, the patient reported symptoms as part of participatory surveillance[1] of acute respiratory infections and submitted a self-collected combined nose and throat swab specimen to the laboratory. On 22 August 2023, the specimen was forwarded at the Dutch National Influenza Centre location at the National Institute for Public Health and the Environment (RIVM) where it tested positive for influenza A virus and negative for A(H1N1)pdm09, both tests included on a commercial 24 pathogen multiplex nucleic acid amplification assay on 23 August.
Further routine subtyping was performed on 24 and 25 August with RT-qPCR assays. Tests for seasonal influenza viruses and H5 viruses were negative. In addition, a laboratory-developed test (LDT) and another commercial assay for generic influenza A virus detection confirmed the presence of type A influenza virus in the specimen.
Routine whole genome sequencing using nanopore technique and virus isolation were started on 28 August. On 30 August, sequencing results revealed that the virus, A/Netherlands/10534/2023, is an A(H1N1)v Eurasian avian-like clade 1C.2.2 swine influenza virus. The HA genome segment clustered closely with recent clade 1C.2.2 swine influenza viruses from 2022 and 2023 from the Netherlands.[2] It clustered less closely with previous clade 1C.2.2 A(H1N1)v (from 2019) and A(H1N2)v (from 2020) viruses from the Netherlands. Phenotypically, the virus is sensitive to neuraminidase inhibitors oseltamivir and zanamivir.
The virus isolate will be shared with the WHO Collaborating Centre in London, United Kingdom of Great Britain and Northern Ireland, and the World Organization for Animal Health (WOAH) avian and swine influenza reference laboratory at the Animal and Plant Health Agency (APHA), UK. The sequences are available from the GISAID database under accession EPI_ISL_18168180.
As of 13 September, the person is recovered. Investigations reported that the individual did not work at a pig farm or other business involving pigs and does not work in health care. Therefore, there is no clear indication of the source of infection.
(SNIP)
WHO risk assessment
Most human cases result from exposure to swine influenza viruses through contact with infected swine or contaminated environments. Occasionally, the source of exposure has remained unidentified.
As these viruses continue to be detected in swine populations worldwide, further human cases following direct or indirect contact with infected swine can be expected.
Current evidence suggests that these viruses have not acquired the ability to sustain transmission among humans. There has been limited, non-sustained human-to-human transmission of variant influenza viruses, although ongoing community transmission has not been identified. In this event, there was no human-to-human transmission resulting in symptomatic disease detected and no further detections have been reported in routine surveillance.
The risk of detecting additional cases associated with this event appears to be low.
If needed, the risk assessment will be reviewed should further epidemiological or virological information become available.