#17,829
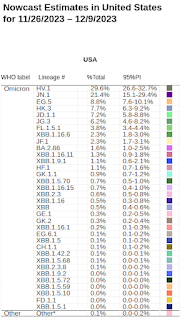
Although we'll get an update tomorrow, in the CDC's most recent Nowcast researchers were actively following nearly 3 dozen variants in the United States, and even more globally. What is important to know is that most are in decline, while a few - particularly JN.1 - are showing strong growth.
Just as Omicron pushed Delta out of the way 2 years ago, a new lineage (BA.2.86 and its descendants, including JN.1) are making life tough for the XBB variants that have reigned for the past year.
While many hoped that acquired `community immunity' would quickly end the COVID threat, viruses haven't survived for hundreds of millions of years on this planet by being so easily defeated.
What we've seen instead is the continued evolution of the SARS-CoV-2 virus, with it becoming more immune evasive over time.
Last summer we began to see reports of a new mutation in the Spike gene (F456L) of several variants increasing their transmissibility (see ECDC Classifies XBB.1.5-like Lineages with the Amino Acid change F456L as Variants of Interest).
In August, we saw the newly emerged and antigenically divergent BA.2.86 variant also carried this F456L mutation. Its direct descendant JN.1 carries it too, along with an additional L455S mutation in the spike protein.
L455F and F456L mutations on the spike protein are often called "FLip" mutations, because they switch the positions of two amino acids, F and L. When you add in the L455S mutation (as with JN.1), they also appear to greatly enhance both transmissibility and immune escape.
As a result, JN.1 is off to the races, and is expected to become the dominant COVID variant in the United States - and around the globe in the weeks ahead.
At least until something `more biologically fit' comes along.
All of which bring us to a rather detailed editorial/review published this week in the journal Viruses, which looks at these new FLip mutations, along with emerging SLIP's and FlipperR's.
Due to its technical nature, I've only posted some excerpts. Those interested in a deeper look will want to follow the link to read it in its entirety. I'll have a brief postscript after the break.The Era of the FLips: How Spike Mutations L455F and F456L (and A475V) Are Shaping SARS-CoV-2 Evolution
by
Daniele Focosi 1,*,Pietro Giorgio Spezia 2,Federico Gueli 3 andFabrizio Maggi 2
Viruses 2024, 16(1), 3; https://doi.org/10.3390/v16010003
Published: 19 December 2023
Convergent evolution of the SARS-CoV-2 Spike protein has been mostly driven by immune escape, in particular by escape to the viral infection-neutralizing antibodies (nAbs) elicited by previous infections and/or vaccinations [1]. These immune escaping mutations usually come at a cost for SARS-CoV-2 fitness, often compromising binding to the ACE2 receptor. So, in order to be competitive, a novel SARS-CoV-2 sublineage has to first largely increase the affinity of the Spike protein for the ACE2 receptor, so that it can later accommodate further immune escape mutations.Recently, the S:F456L mutation has been convergently acquired by many independent XBB.1* sublineages and by a single BA.2.75.3-derived sublineage (DV.7.1), soon followed by the S:L455F mutation, a phenomenon referred to as “FLip” (from the initials of the mutated amino acid residues).The acquisition has sometimes been detected as simultaneous (see the lower part of Figure 1), but it should be considered that low genomic surveillance rates in 2023 could account for missing the intermediate step. When the parent lineage also shows the S:K478R mutation—either occurring before the “FLip” (as in JF.1), acquired simultaneously with the “FLip” (as in GW.5), or developed after the “FLip” (as in GK.1.4 and in JR.1.1)—the lineage instead comes under the nickname “FLippeR”.Real world data have shown that lineages which were outcompeted and declining started regrowing in a sustained manner after gaining the FLip, such as XBB.1.5-derived XBB.1.5.70* and JD.1* or CH.1.1-derived DV.7.1. The biological characteristics of one of the FLips, namely HK.3, have recently been reported in detail [2].
Figure 1. Summary of PANGOLIN-designated SARS-CoV-2 Omicron sublineages which have acquired the S:F456L mutation as of 18 December 2023.
Both S:F456L and S:L455F largely increase ACE2 affinity and their combination is synergic [3]. S:L455F also confers some immune escape [4] (and had been observed to emerge after treatment with casirivimab [5]), but FLip lineages remain sensitive to class 1 antibodies [3]. This has also been proven true for BA.2.86 (Pirola) [6], which soon gained fitness in the S:L455S-positive descendant JN.1 [7,8]. Amazingly, JN.1 sequences which have later acquired F456L (nicknamed “Slip’s”) were reported in late November 2023 from France and New Zealand.
(SNIP)
In conclusion, SARS-CoV-2 is again confirming its incredible plasticity in escaping the consolidating human immune response. Since S:F456L, S:L455F, and S:A475V do not occur in the recently marketed XBB.1.5-based “updated” vaccines, the extent to which nAbs in vaccine recipients will provide protection from severe disease remains to be established. Epidemiological monitoring is highly recommended to assess the relationships between specific sublineages and increased clinical severity.
While a lot of people hoped that within 12 to 18 months we'd all either have contracted the virus - or have been vaccinated - and that we'd have acquired `herd immunity', after 4 years the virus continues to reinvent itself in new and unexpected ways.
Although it is possible the SARS-CoV-2 virus will eventually run out of `biologically fit' avenues for immune escape, for now, the virus appears to have a new set of tools to play with going into 2024.