One of the biggest problems when trying to assess - and then communicate - the risks from HPAI H5 is that we are not dealing with a single entity; but rather a growing array of continually evolving viruses which are spreading through hundreds of avian and mammalian species around the globe.
The H5N1 virus spreading in cattle (genotype B3.13) is genetically distinct from the H5N1 virus found spreading in mink in Spain, or the H5N1 virus killing marine mammals in South America, or the H5N5 virus in raccoons in Nova Scotia,
There are literally hundreds of H5 variants circulating around the globe - many of which aren't even on our radar - with more being added with each passing day. While most will end up evolutionary failures, some will be more adept at infecting, spreading, or causing disease in mammals than others.
We know some (but not all) of the amino acid changes that suggest mammalian adaptation, and so when we see those crop up, we take notice. But global surveillance and reporting are far from robust, and we are likely only seeing the tip of the iceberg.
With all of this in mind, today we have a study that looks at how a specific variant of H5N1 - one that raised concerns after it was found spreading efficiently in mink in Spain in November of 2022 - infects, replicates, and spreads in pigs.
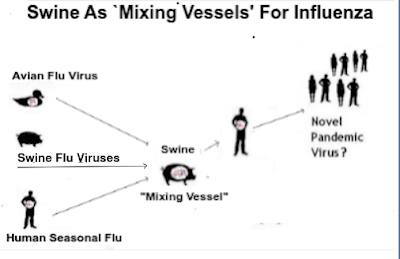
Pigs are a particular concern because they are susceptible to a wide variety of human, swine, and avian flu viruses, making them likely `mixing vessels', where reassortment may occur.
We've seen some anecdotal reports of HPAI H5 in pigs over the years, including:
WHO H5N1 detected in pigs in China (2004)
An Unusual Report Of H5N1 in Pigs (Indonesia 2016)
But past attempts to infect pigs experimentally have suggested pigs may not be an ideal host for H5N1 (see 2023's EID Journal: Low Susceptibility of Pigs against Experimental Infection with HPAI Virus H5N1 Clade 2.3.4.4b).
That study came with a significant caveat:
However, considering the ongoing massive panzootic of this virus, a plethora of new genotypes of the circulating strain is emerging, with possibly higher permissiveness for pigs.
What might happen over time, outside of a controlled laboratory environment, is not known.
Due to its length, and technical nature, I've only posted the link, abstract, and some excerpts from the study. Follow the link to read it in its entirety. I'll have a bit more after the break.
Research Article
Taeyong Kwon,Jessie D. Trujillo,Mariano Carossino,Eu Lim Lyoo,Chester D. McDowell,Konner Cool,
Article: 2353292 | Accepted author version posted online: 07 May 2024
https://doi.org/10.1080/22221751.2024.2353292Abstract
Rapid evolution of highly pathogenic avian influenza viruses (HPAIVs) is driven by antigenic drift but also by reassortment, which might result in robust replication in and transmission to mammals. Recently, spillover of clade 2.3.4.4b HPAIV to mammals including humans, and their transmission between mammal species has been reported. This study aimed to evaluate the pathogenicity and transmissibility of a mink-derived clade 2.3.4.4b H5N1 HPAIV isolate from Spain in pigs.
Experimental infection caused interstitial pneumonia with necrotizing bronchiolitis with high titers of virus present in the lower respiratory tract and 100% seroconversion. Infected pigs shed limited amount of virus, and importantly, there was no transmission to contact pigs.
Notably, critical mammalian-like mutations such as PB2-E627K and HA-Q222L emerged at low frequencies in principal-infected pigs. It is concluded that pigs are highly susceptible to infection with the mink-derived clade 2.3.4.4b H5N1 HPAIV and provide a favorable environment for HPAIV to acquire mammalian-like adaptations.
Discussion
(Excerpts)
One of key findings in this study was that experimental infection of pigs with the mink-derived clade 2.3.4.4b H5N1 virus resulted in productive virus replication and seroconversion in 100% of the principal-infected pigs. Virus titers in BALF samples of principal-infected animals reached 103.3 to 104.7 TCID50/mL at 3 DPC and declined to 102.7 to 103.5 TCID50/mL at 5 DPC.
In addition, the virus was found in other respiratory tissues, such as trachea, bronchi, as well as lymphoid tissues, such as tracheobronchial lymph nodes and tonsils. Moreover, gross and histological evaluation of various tissues demonstrated that the infection caused acute multifocal to coalescing necrotizing broncho-interstitial pneumonia at 3 and 5 DPC, with residual mild interstitial pneumonia still present at 21 DPC.
(SNIP)
Experimental infection of pigs with LPAI viruses without mammalian-like adaptations were also previously performed, but none of the experimental infections with LPAI H5 viruses resulted in a productive infection [36].
(SNIP)
One of the important findings is that H5N1 viruses inoculated into pigs acquired mammalian-like mutations/adaptations, which potentially results in increased fitness of the H5N1 virus in mammalian hosts. There is a total of four mink-derived H5N1 isolates from the infected Spanish farm which were sequenced; all of them possessed the PB2-T271A mutation. The PB2-T271A mutation is known to lead to enhanced polymerase activity in mammalian cells and might contribute to cross-species transmission from avian species to mammals including mink [13,44]. Our results show that the PB2-T271A mutation was stable with no evidence of a change in the frequency of the mutation after replication of the virus for several days in pigs. However, minor variants possessing the mammalian-like E627K mutation in PB2 emerged at low frequencies in this study.
The PB2-E627K mutation is a key determinant for mammalian adaptation of avian influenza viruses; its introduction into the genome of an avian IAV enables efficient replication of the avian-origin polymerase complex in mammalian cells; it increases virulence in mice, and contributes to air-borne transmission in ferrets and contact transmission in guinea pigs [45-50]. Similarly, the PB2-E627V substitution was shown to increase viral replication in mammalian cells and virulence in mice models [51]; it was found in clinical samples in our study as a minor virus population. Furthermore, one oropharyngeal swab possessed a PB2-K526R mutation at the frequency of 3.1%; the K526R mutation was shown to enhance the effect of the PB2-E627K on avian influenza virus replication in mammalian cells and also their virulence in mice [52].
Another key mutation is the HA-Q222L (according to the H5 numbering system) which is equivalent to a Q226L mutation using the H3 numbering system. This mutation was found in one oropharyngeal swab at 1 DPC. Several amino acids in the HA receptor binding site are responsible for switching host specificity; however, the Q222L and G224S substitutions in the H5 numbering (Q226L and G228S in the H3 numbering) are crucial for increased mammalian-like α2,6-sialic acid receptor preference in different subtypes of avian IAVs; they are located in the receptor binding domain and interact directly with the host’s sialic acid receptor [53]. In addition, the HA-Q222L mutation was previously described to contribute to the emergence of transmissible H5N1 viruses via aerosols after serial passage of H5N1 in ferrets [47,48]. Despite the critical role of these mutations in pathogenicity and transmissibility of IAVs in mammalian hosts, it still remains unclear how the H5N1 clade 2.3.4.4b virus underwent its molecular evolution, and eventually acquired critical mammalian-like mutations.
In addition to the role of pigs in the ecology of IAVs as the “mixing vessel”, the emergence of mammalian-adapted H5N1 viruses in pigs is close to reality; therefore, pigs may serve as the “hotspot” to introduce mammalian-like mutations into avian IAVs although the frequencies of the mammalian-like mutations described here are rather low.
Spillover of H5N1 HPAI clade 2.3.4.4b viruses to mammals and their sustained transmission between mammals has been reported [13,54]. The majority of mammals infected by the HPAI H5N1 clade 2.3.4.4b virus are carnivores, and the consumption of infected dead wild birds is presumably the basis for these spillover events [55]. In contrast to the HPAI H5N1 clade 2.3.4.4b virus infections in wild avians and wild mammals, outbreaks in farmed avian and mammal species pose a much greater risk to humans, due to its close proximity to occupational workers and -in the case of mammalian species - the favorable environment for the acquisition of mammalian-like adaptations.
Therefore, evaluating the infectivity/susceptibility and transmissibility of the emerging HPAI viruses in well-established mammalian animal models such as swine and ferrets is crucial to determine their pandemic potential.
The present study demonstrated that infection of pigs with the mink-derived clade 2.3.4.4b H5N1 virus led to a productive viral replication in the respiratory tracts of all principal-infected pigs including seroconversion; however, virus shedding from principal-infected pigs was not high and/or frequent enough for an efficient transmission to co-mingled sentinel pigs.
Notably, key mammalian-like mutations were found in samples derived from principal-infected pigs. In conclusion, the results obtained in the present study suggest that the mink-derived H5N1 clade 2.3.4.4b virus exhibited increased infectivity in pigs when compared to avian-origin H5N1 clade 2.3.4.4b viruses, however, did not transmit to co-mingled sentinel animals; therefore, this virus would still be placed in a moderate risk group in terms of transmission ability to humans.
The very good news is this particular mink-derived H5N1 virus still doesn't transmit efficiently among co-housed pigs, but it is significantly more pathogenic in swine than past studies have suggested.
While still not ready for prime time, HPAI H5 continues to make progress in adapting to mammals.
Good research takes time and resources, which is why we are looking at the abilities of a 2022 mink-derived virus rather than the current cattle B3.13 genotype in pigs. Hopefully those studies are being fast-tracked.
While there are a lot of pathways that HPAI H5 could take to becoming a pandemic, the `easiest' route may be through pigs.
In swine the virus would have ample opportunities to reassort with already `humanized' H1, H2, and H3 viruses as well as frequent contact with humans. Which is why increasing biosecurity for pigs needs to be high on our list of priorities.
And something we do before it becomes a fait accompli.