# 8766
Although influenza A viruses are believed to have originated in avian species (primarily waterfowl), over time they’ve managed to infect, and adapt to, a wide variety of mammals. Humans, obviously, and swine . . . but we’ve also seen `human’ flu viruses documented in turkeys, skunks, ferrets, cats, elephant seals and dogs.
In That Touch Of Mink Flu I wrote about 11 farms in Holstebro, Denmark that were reported to be infected with a variant of the human H3N2 virus.
Less commonly reported - camels, whales and seals have all been shown to be susceptible to influenza viruses (cite Evolution and ecology of influenza A viruses R.G. Webster et al.) In 2012, an avian flu strain was identified in New England seals (see mBio: A Mammalian Adapted H3N8 In Seals).
While influenza viruses are generally categorized as being avian, swine, human, or equine – their host range is far greater than that.
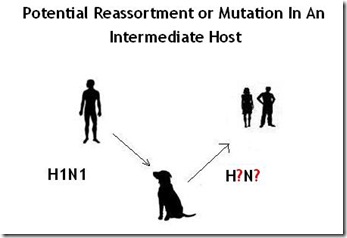
And about a decade ago, we saw that host range formally expand to canines as well.
In 2004, the H3N8 equine influenza – a strain that has been around nearly a half century – suddenly jumped, and adapted to dogs, creating a new dog-specific (canine) lineage of H3N8. Since then, H3N8 has continued to spread among dogs, although it has never been known to infect humans.
But, as noted above, on rare occasions humanized influenza viruses have been shown to infect dogs as well, along with avian flu strains like H5N1 and H5N8 (see Korea Finds More Dogs With H5N8 Antibodies).
This ability to be infected by more than one type of influenza virus opens the door to the possibility that dogs could serve as a `mixing vessel’ for influenza viruses, through a process called Reassortment.

Reassortment, or `Shift’ occurs when a single host is simultaneously infected with two different influenza viruses. When that happens, it is possible for viruses to swap parts of their genetic sequence, and create a new, hybrid virus.
While most of the time, this leads to an evolutionary failure, every once in awhile this process produces a viable, even highly competitive, new flu strain.
It was a series of reassortments in swine, over a period of years, that led to the creation of the 2009 H1N1 pandemic virus, and the recent outbreak of H7N9 in China, and the H5N8 outbreaks in Korea, are apparently due to viral reassortment in avian species.
We normally think of birds, or swine, as prime `mixing vessels’ for influenza, but the truth is, any host that can support dual infection has the potential to produce a `hybrid’ flu virus.
In the past we’ve looked at a number of species with at least theoretical potential to act as mixing vessels.
Mixing Vessels For Influenza
A Host Of Reservoirs
Virology Journal: Receptor Cells In Minor Poultry Species
The (Swine) Influenza Reassortment Puzzle
All of which serves as prelude to a new study, which appears ahead of print in the Journal of Virology, that looks at the ability of different influenza strains to infect, and replicate in, canine tracheal tissues.
First a link to the study (PDF), a link to the abstract, followed by excerpts from a press release from the American Society for Microbiology, and then some closing comments.
Infection and pathogenesis of canine, equine and human influenza viruses in canine tracheas.
Gaelle Gonzaleza, John F. Marshallb, Joanna Morrella, David Robbc, John W. McCauleyd, Daniel R. Pereze, Colin R. Parrishf and Pablo R. Murciaa#
ABSTRACT
Influenza A viruses (IAVs) can jump species barriers and occasionally cause epidemics, epizootics, pandemics and panzootics. Characterizing the infection dynamics at the target tissues of natural hosts is central to understanding the mechanisms that control host range, tropism and virulence. Canine influenza virus (CIV, H3N8) originated after the transfer of an equine influenza virus (EIV) into dogs. Thus comparing CIV and EIV isolates provides an opportunity to study the determinants of influenza emergence.
Here we characterize the replication of canine, equine and human IAVs in the trachea of the dog, a species to which humans are heavily exposed.
We defined a phenotype of infection for CIV, which is characterized by high levels of virus replication and extensive tissue damage. CIV was compared to evolutionary distinct EIVs, and the early EIV isolates showed an impaired ability to infect dog tracheas, while EIVs that circulated near the time of CIV emergence exhibited a CIV-like infection phenotype. Inoculating dog tracheas with various human IAVs (hIAVs) showed that those infected the tracheal epithelium with various efficiencies depending on the virus tested. Finally, we show that reassortant viruses carrying gene segments of CIV and hIAV are viable and that addition of the hemagglutinin (HA) and neuraminidase (NA) of CIV to the 2009 human pandemic virus results in a virus that replicates at high levels and causes significant lesions. This provides important insights into the role of evolution on viral emergence and on the role of HA and NA as determinants of pathogenicity.
(Continue . . . )
This from the American Society for Microbiology.
Evolution of Equine Influenza Led to Canine Offshoot Which Could Mix With Human Influenza
CONTACT: Jim Sliwa
jsliwa@asmusa.org
WASHINGTON, DC – June 19, 2014 – Equine influenza viruses from the early 2000s can easily infect the respiratory tracts of dogs, while those from the 1960s are only barely able to, according to research published ahead of print in the Journal of Virology. The research also suggests that canine and human influenza viruses can mix, and generate new influenza viruses.
Canine influenza is a relatively new disease. The first appearance is believed to be in 2003, as a result of direct transfer of a single equine influenza virus to dogs in a large greyhound training facility and was subsequently carried to many states by the infected greyhound, say the researchers. Similar transfers have occurred among foxhounds in the UK, and in dogs kept near infected horses during a 2007 outbreak in Australia, they report.
In the study, investigators from the United States and the United Kingdom infected dog tracheal explant cultures—essentially pieces of trachea cultured in the laboratory to mimic the cellular complexity and the host physiology of the host—with canine influenza virus, equine influenza virus, and human influenza viruses. (The use of explants has increased in recent years, and they have been shown to be useful for studies of viral pathogenesis.) They then compared the growth of the viruses, and the damage they wrought.
Worthy of note, in Korea a different canine influenza virus emerged in 2007. Unlike the canine H3N8 virus seen in the United States, this canine H3N2 appears to have jumped directly from an avian source. In 2008 the CDC’s EID Journal carried the following report on this newly emerging canine flu:
Transmission of Avian Influenza Virus (H3N2) to Dogs
In 2011, we saw the plot thicken yet again, when it was announced that this canine H3N2 had jumped to cats (see Korea: Interspecies Transmission of Canine H3N2). In their abstract, the authors wrote:
Our study for the first time shows that cats are susceptible to canine influenza H3N2 infection, suggesting that cats may play an intermediate host role in transmitting the H3N2 virus among feline and canine species, which could lead to the endemic establishment of the virus in companion animals.
Such a scenario raises a public health concern, as the possibility of the emergence of new recombinant feline or canine influenza viruses in companion animals with the potential to act as zoonotic infection cannot be excluded.
While pigs and birds may be far better `mixing vessels’ for influenza than dogs or cats will ever be, companion animals are of particular concern because of how closely humans interact with them.
All of this just shows how remarkable influenza viruses are at adapting to new hosts, and re-inventing themselves.
And why we ignore their progress in the wild at our own peril.