#17,871
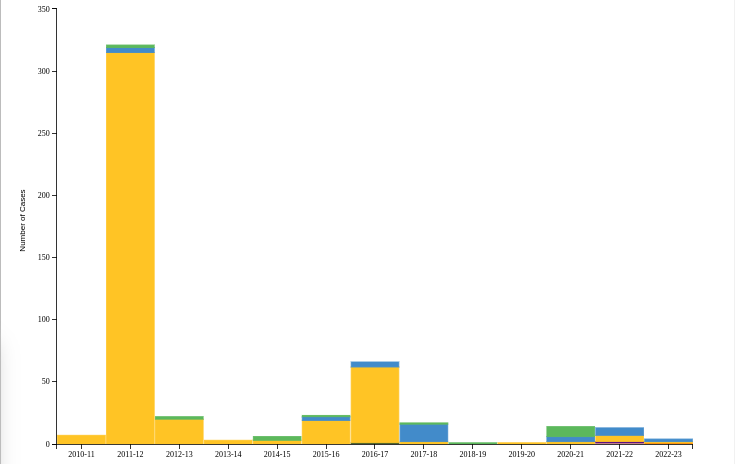
Although many cases likely go undetected - every year we see sporadic detections of swine variant (H1N1v, H1N2v, H3N2v) infection in humans here in the United States (see CDC chart below) and around the world.Last November the UK reported their first laboratory-confirmed swine variant H1N2v infection (see UKHSA Identifies 1st H1N2v (Swine Variant) Infection In the UK), which raised alarm bells and set off an immediate public health investigation.
Throughout December we followed reports from the UKHSA (see UKHSA Publishes Update & Risk Assessment On H1N2v), including the release of some stringent isolation and testing protocols (see UKHSA Issues Guidance For Public On Possible H1N2v Exposure).
Yesterday the ECDC's journal Eurosurveillance published a Rapid Communications on this event, which describes 1 laboratory confirmed - and two probable - infections that were detected by the UK's epidemiological investigation.
While many swine variant cases report recent contact with pigs, this recently (seasonal flu) vaccinated index case had no direct contact with pigs, and so their source of infection remains unknown.
Due to its length, I've only posted some excerpts, so follow the link to read the report in its entirety. I'll have a bit more after the break.
A case of swine influenza A(H1N2)v in England, November 2023
Jade Cogdale1 , Beatrix Kele1 , Richard Myers1 , Ruth Harvey2 , Abi Lofts2 , Tanya Mikaiel2 , Katja Hoschler1 , Ashley C Banyard3 , Joe James3 , Benjamin C Mollett3,4 , Alexander MP Byrne3 , Jamie Lopez-Bernal1 , Conall H Watson1 , Meera Chand1 , William Welfare5 , Deborah A Williamson1 , Isabel Oliver1 , Simon Padfield6 , Andrew Lee6 , Suzanne Calvert6 , Martin A Bewley6 , Louise Wallace7 , Simon deLusignan8,9 , Nicola S Lewis2,4 , Ian H Brown3 , Maria Zambon1 , on behalf of the Influenza A(H1N2)v Incident Management Team1
Case detection and description
The United Kingdom Health Security Agency (UKHSA) operates community respiratory virus surveillance with the Royal College of General Practitioners (RCGP) Research and Surveillance Centre (RSC) [1,2]. In November 2023, an influenza A-positive sample from the RCGP surveillance scheme was confirmed as subtype H1N2. The sample was taken from a an 80-year-old individual in northern England who presented with a 4-day history of cough, shortness of breath and production of green sputum. The individual had been vaccinated 6 weeks prior with 2023/24 seasonal influenza vaccine and was treated with oral antibiotic (amoxicillin) but did not require hospitalisation. The illness gradually resolved over the following week.
Initial testing of the respiratory sample indicated an influenza A infection with high quantification cycle value (Cq = 33), close to the normal limit of detection (Cq = 40). The subtype of the sample was not identified by RT-PCR for detection of seasonal influenza A (H1N1)pdm09 or H3, suggesting the possibility of an unusual virus variant. Whole genome sequence analysis undertaken using the Illumina platform (Illumina, San Diego, United States) was consistent with a swine H1N2 virus infection belonging to clade 1B.1.1 [3,4].
(SNIP)
Comprehensive backward contact tracing from the index case identified that, although the individual lived in a region of the country with a high density of pigs, there was no animal contact in the history of the case or their household contact (no pigs, no pets and no contact with environments that were obviously contaminated by animals).
A household contact had also been unwell around the same time but had not sought medical attention or been swabbed. For both individuals, the illness resolved over a period of 7–10 days. The household contact who developed clinical disease was therefore classified as a probable case.
On forward contact tracing, four additional individuals were identified, three of whom were asymptomatic following potential exposure (to the index case) but had swabs taken on a precautionary basis. None of them tested positive for influenza A. The fourth contact was a healthcare worker (HCW) who had seen the index case at an outpatient appointment on the day after illness onset regarding an unrelated condition. The HCW became unwell with a mild respiratory illness involving a runny nose and cough 9 days after exposure to the index case and was designated as a further probable case. The illness was considered minor and resolved fully without complications. A swab taken on 26 November (10 days after symptom onset) tested negative for influenza A but positive for rhinovirus.
Overall, we identified seven individuals, of whom two were designated as probable cases. Secondary contact tracing was initiated around the HCW probable case on a precautionary basis, and 198 contacts (38) with symptoms compatible with influenza-like illness (ILI) were identified. For 149 of them, PCR tests have been requested, with 88 negative results for influenza A, one positive for H1N1 and 60 results pending or lost to follow-up at 17 January 2024.
(SNIP)
Viral factors permitting swine-to-human zoonotic infection remain largely undefined [11]. It is currently not possible to predict which of the clades of swine influenza A viruses causes human infections or has a propensity for sustained human-to-human transmission, leading to a global pandemic such as occurred in 2009. The risk of variant infection of humans relates to the risk of mucosal contamination and aerosols at animal–human interfaces such as, but not limited to, farm environments, live animal markets and agricultural fairs, along with the nature of the variant virus and unknown virus and host characteristics.
To ensure preparedness, WHO Human seasonal influenza Vaccine Composition Meetings also include the review of swine candidate vaccine strains (CVVs), updated regularly to be representative of circulating swine strains [5]. Based on the assessments undertaken following this case detection, currently available CVVs for H1 1B swine influenza viruses are unlikely to afford protection against the H1 1B.1.1 swine influenza A viruses detected in GB.
Conclusion
The identification of this variant infection occurred due to an unusual pattern of reactivity in laboratory diagnostic assays. Work is underway with the Medicines and Regulatory Agency, to assess the detection capability of-commercial and non-commercial platforms used in the United Kingdom. Gaining assurance about the ability of available clinical diagnostic assays to detect swine variant influenza viruses is a complex task, with the necessity to develop rapid response arrangements for clinical diagnostic assurance when new influenza variants are detected in humans, and to develop external quality assessment schemes with a broad range of zoonotic viruses.
While the UK maintains enhanced surveillance for novel flu viruses due to the presence of H5N1 (see UK HSA Guidance: Investigation & Initial Clinical Management Of Possible Human Avian Flu Infection), the detection of this index case (and the two probables) was largely a matter of luck.
Not everyone who gets the `flu' goes to their GP, and only a subset of those have a sample taken and sent off for RT-PCR testing. Even when both happen, there is a relatively short `window' for accurate sample collection.What is GP surveillance data?
A third of GPs in England opt to share pseudonymised patient data with the Oxford Royal College of GPs Research and Surveillance Centre (RSC). Some also take swabs and samples – virology swabs are sent to the UK Health Security Agency (UKHSA) to monitor infections and vaccine effectiveness, and serology samples are used to report background levels of natural immunity and measure vaccine waning.
(SNIP)
In November 2023, the first confirmed case of Influenza A(H1N2)v in humans in the UK, unofficially referred to by the media as "swine flu", was detected through routine national flu surveillance undertaken by the network.
In order to detect this case, public health had to get `lucky' 3 times.
Last spring, a study from the UK HSA (see UK Novel Flu Surveillance: Quantifying TTD) estimated the TTD (Time To Detect) a novel H5N1 virus in the community via passive surveillance could take weeks, and the virus might only be picked up after hundreds or possibly even thousands of infections.
Many countries have neither the ability, or inclination, to conduct this level of surveillance.
Results. We estimate that the median multiplier for children was 200 (90% range, 115–369) and for adults was 255 (90% range, 152–479) and that 2055 (90% range, 1187–3800) illnesses from H3N2v virus infections may have occurred from August 2011 to April 2012, suggesting that the new virus was more widespread than previously thought.
And while the origins of the SARS-COV-2 virus remain shrouded in mystery, there is evidence that suggests it was circulating in humans for months before it appeared in Wuhan, China in late 2019.
It is likely that many `mini-outbreaks' of zoonotic viruses occur each year that fly under our radar. They emerge, spread briefly, but are not biologically `fit' enough to thrive. Some will never make the grade, but others (like HPAI, swine flu, MERS-CoV, etc.) may only need an evolutionary nudge or two.
Probably the only thing we can say with any level of certainty is, we probably won't see it until it is already well-established, and `biologically' ready for prime time.