#18,802
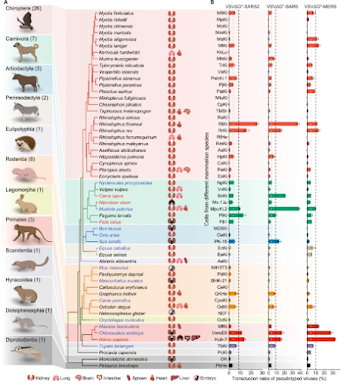
While exquisitely adapted to humans, the highly mutable SARS-CoV-2 virus has found a home in a wide range of other mammalian species, including mink, deer, cats, and even rodents (see Nature: Comparative Susceptibility of SARS-CoV-2, SARS-CoV, and MERS-CoV Across Mammals).
In late 2020 Danish authorities were alarmed to discover several mutated strains of COVID-19 had arisen in farmed mink and had spilled-back repeatedly to humans (see Emergence and spread of SARS-CoV-2 variants from farmed mink to humans and back during the epidemic in Denmark, June-November 2020).
Although that emergency was relatively short-lived (the aggressive Alpha COVID variant emerged in late 2020 and quickly supplanted these mink-variants), it did illustrate the problem; carriage of SARS-CoV-2 by other host species can produce new variants, some of which can potentially jump back into humans.
While controversial, there is even some evidence to suggest that the Omicron variant may have evolved after the SARS-CoV-2 virus jumped to mice or other rodents (see Evidence for a mouse origin of the SARS-CoV-2 Omicron variant), and then spilled back into humans.
We've also seen evidence that older COVID variants - no longer circulating in humans - can be maintained long-term in non-human host populations (see PNAS: White-Tailed Deer as a Wildlife Reservoir for Nearly Extinct SARS-CoV-2 Variants), and may then follow new, and divergent evolutionary paths.
And while rarely reported, reports of suspected pet-to-human transmission have also been documented (Journal: Suspected Cat-to-Human Transmission of SARS-CoV-2 - Thailand).
Spillovers into farmed animals, however, are particularly worrisome, because they allow for serial transmission across a large number of hosts, which may result in host adaptation.

But the concern isn't just over COVID.
Two summers ago, in PNAS: Mink Farming Poses Risks for Future Viral Pandemics, we looked at an opinion piece by Professor Wendy Barclay & Tom Peacock on why fur farms - and mink farms in particular - are high risk venues for both flu and SARS.Fur farms, very much like live poultry (and bush meat) markets, pose significant risks of harboring and spreading avian flu, novel coronaviruses, and other zoonotic diseases (e.g. Ebola, Lassa Fever, SFTS, Anthrax, etc.).
Despite these repeated warning signs, surveillance and testing of farmed animals for novel viruses remains quite limited. Some countries have made attempts to close, or better regulate, fur farms and live markets, but many operate with few if any safeguards.
All of which bring us to a recent preprint, which looks at the unchecked, long-term spread of multiple COVID variants within and between Lithuanian mink farms, and evidence of repeated human-to-mink, and mink-to-human, transmission.
We analysed 1 323 SARS-CoV-2 genomes from humans and 58 from mink to assess the effectiveness of Lithuania's genomic surveillance programme and to evaluate the risks associated with mink farming.
We identified mink-associated human infections bearing mutations associated with mink adaptation, commonly affecting mink farm employees and caused by SARS-CoV-2 lineages that have been extinct in the general human population for months
Beyond the hard science presented (albeit based on limited data), this report is an excellent reminder of the risks posed by fur farming and along with the risks of taking a passive approach to monitoring and containing zoonotic diseases in livestock.
An approach that isn't limited to fur farms (see FAO: Recommendations for the Surveillance of Influenza A(H5N1) in cattle).
Martynas Smičius, Ingrida Olendraitė, Jonas Bačelis,Aistis Šimaitis, Miglė Gabrielaitė, Bas B Oude Munnink, Reina S Sikkema, Arūnas Stankevičius, Žygimantas Janeliūnas, Paulius Bušauskas, Egidijus Pumputis, Simona Pilevičienė, Petras Mačiulskis, Marius Masiulis, Vidmantas Paulauskas, Snieguolė Ščeponavičienė, Monika Katėnaitė, Rimvydas Norvilas, Ligita Raugienė, Rimvydas Jonikas, Inga Nasvytienė, Živilė Žemeckienė,Kamilė Tamušauskaitė, Milda Norkienė, Emilija Vasiliūnaitė, Danguolė Žiogienė, Albertas Timinskas, Marius Šukys, Mantas Šarauskas, ProfileDovilė Juozapaitė,Daniel Naumovas, Arnoldas Pautienius, Astra Vitkauskienė, ProfileRasa Ugenskienė, Alma Gedvilaitė, Darius Čereškevičius, Laimonas Griškevičius, Marion Koopmans, Alvydas Malakauskas, Gytis Dudas
AbstractSeveral studies have documented reverse zoonotic transmission of SARS-CoV-2, including in farmed mink which are susceptible to human respiratory viruses and are known for serving as a reservoir capable of generating new virus variants in densely populated farms.Here, we present the results of a genomic investigation launched in response to detected human infections with mink-origin SARS-CoV-2 lineages, and show evidence of at least 14 high-confidence introductions of SARS-CoV-2 from humans into farmed mink in Lithuania where sustained transmission in farmed mink lasted up to a year.
We estimated the most likely timeframes for these introductions encompassing at least six SARS-CoV-2 lineages, some of which were already extinct in humans, with Bayesian phylogenetic and molecular clock analyses.This study highlights the public health risks posed by fur farms and underscores that passive genomic surveillance systems are ineffective without the active involvement and expertise of responsible institutions.
Surveillance gaps in Lithuania
Our analysis revealed multiple likely human-to-mink SARS-CoV-2 jumps during the COVID-19 pandemic, some of which circulated undetected within mink populations for extended periods and resulted in anthropozoonotic spillovers back to humans.
Essentially, our findings highlight critical gaps in the passive surveillance (especially genomic) system, hindered by the absence of incentives for self-reporting and insufficient communication between human and animal health institutions.
Ultimately, the lack of decisive action from institutions responsible for animal health can pose a serious threat to human health and addressing these issues is crucial to improve preparedness and response during zoonotic outbreaks in future pandemics.
Admittedly, dozens of other countries reported similar contemporaneous outbreaks, but individual national responses differed greatly. Denmark and the Netherlands quickly ordered mass cullings (see Denmark Orders Culling Of All Mink Following Discovery Of Mutated Coronavirus), while some countries tried far more conservative approaches.
Some countries kept the details of their responses pretty close to their vest (e.g. Poland & Finland), while others have remained silent (e.g. China, North Korea, Russia).
The fur trade remains a significant part of many country's economies, and the fur lobby still carries considerable weight.
That said, there is a growing movement to ban fur farms, particularly in the EU. In September 2023, Lithuania became the 20th European country to completely ban fur farming, although this is still being phased in and won't come into full effect until Jan 1st, 2027.
Poland and Russia remain major fur producers, along with the United States and Canada. China, however, remains the largest global producer and exporter of farmed fur, with 2/3rds of the world's mink farms located there.Estonia, Romania, and Latvia have authorized bans that will begin between 2026-2028. Some countries have enacted partial bans, while others continue to debate the matter.
Reportedly fox and raccoon dogs are raised on many of these same farms, increasing the odds of inter-species transmission of novel viruses.
Worse, raw poultry or poultry meat products are sometimes fed to mink/foxes raised in captivity, raising concerns that they could become infected with one of the many avian flu subtypes found in Asian poultry.
While the next pandemic virus could certainly emerge directly from the wild, the practice of raising millions of highly (flu & COVID) susceptible animals in densely populated farms greatly increases that pandemic threat.
Whether we can summon the political will to take the necessary steps needed to prevent that from happening, remains to be seen.
But time is definitely not on our side.