#17,251
While SARS-CoV-2 has become extraordinarily well-adapted to humans, it has also demonstrated the ability to infect other mammals, raising concerns that it could follow alternate evolutionary paths that could lead to the future spillover of new, potentially more problematic, variants.
In the fall of 2020 SARS-CoV-2 jumped from humans to farmed mink in Denmark, and began to mutate into new mink-variants (see Denmark Orders Culling Of All Mink Following Discovery Of Mutated Coronavirus), resulting in several mutated viruses that jumped back into humans (see WHO 2nd Update: SARS-CoV-2 mink-associated variant strain – Denmark).It is not an idle concern, as we've already seen it happen.
Similar events have been reported in the United States (see CDC: Investigating Possible Mink-To-Human Transmission Of SARS-CoV-2 In The United States) and in Hong Kong (see Hong Kong Detects COVID In Pet Store Hamsters - Suspends Sales & Orders Cull).
And while controversial, there is even some evidence to suggest that the Omicron variant may have evolved after the virus jumped to mice or other rodents (see Evidence for a mouse origin of the SARS-CoV-2 Omicron variant), and then spilled back into humans (for more, see Maryn McKenna's Wired article Where Did Omicron Come From? Maybe Its First Host Was Mice).
The threat of new COVID variants emerging from non-human hosts is demonstrably real, and we ignore them at our considerable peril.
Although we have one-off reports from around the world, and the USDA maintains a dashboard of mammals infected with COVID in the United States (see below), there is still a lot we don't know about the host range of the SARS-CoV-2 (and SARS-CoV & MERS-CoV) viruses.
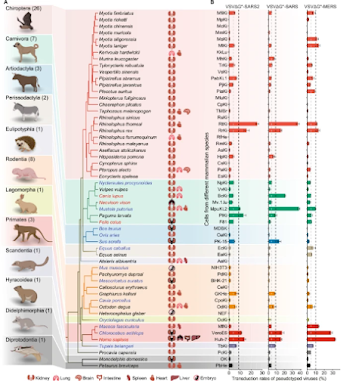
All of which brings us to a lengthy, highly detailed (and often technical) open-access study published yesterday in the Journal Nature, that provides a comparative in vitro infection analysis of 83 cell cultures from 55 species of mammals, using pseudotyped viruses with spike proteins from the original SARS-CoV, MERS-CoV, and SARS-CoV-2.
While not the perfect analog for infection in the wild, in vitro studies using pseudotyped viruses are the only practical way to test the susceptibility of large numbers of species. With more than 5,400 species of mammals on this earth, this study can't tell us the whole story, but it is an excellent start.
Due to its length, I'm only going to post some excerpts from the Abstract and Discussion, but I would encourage you to follow the link to read the full report.
Comparative susceptibility of SARS-CoV-2, SARS-CoV, and MERS-CoV across mammalsMeng Li,Juan Du, Weiqiang Liu, Zihao Li, Fei Lv, Chunyan Hu, Yichen Dai, Xiaoxiao Zhang, Zhan Zhang, Gaoming Liu, Qi Pan, Yang Yu, Xiao Wang, Pingfen Zhu, Xu Tan, Paul A. Garber &
Xuming ZhouAbstract
Exploring wild reservoirs of pathogenic viruses is critical for their long-term control and for predicting future pandemic scenarios. Here, a comparative in vitro infection analysis was first performed on 83 cell cultures derived from 55 mammalian species using pseudotyped viruses bearing S proteins from SARS-CoV-2, SARS-CoV, and MERS-CoV.Cell cultures from Thomas’s horseshoe bats, king horseshoe bats, green monkeys, and ferrets were found to be highly susceptible to SARS-CoV-2, SARS-CoV, and MERS-CoV pseudotyped viruses.Moreover, five variants (del69-70, D80Y, S98F, T572I, and Q675H), that beside spike receptor-binding domain can significantly alter the host tropism of SARS-CoV-2.An examination of phylogenetic signals of transduction rates revealed that closely related taxa generally have similar susceptibility to MERS-CoV but not to SARS-CoV and SARS-CoV-2 pseudotyped viruses.Additionally, we discovered that the expression of 95 genes, e.g., PZDK1 and APOBEC3, were commonly associated with the transduction rates of SARS-CoV, MERS-CoV, and SARS-CoV-2 pseudotyped viruses.This study provides basic documentation of the susceptibility, variants, and molecules that underlie the cross-species transmission of these coronaviruses.(SNIP)
Discussion
Herein, we have demonstrated that SARS-CoV-2, SARS-CoV, and MERS-CoV can infect the cells of dozens of mammal species, indicating they are generalist viruses and not specifically adapted to humans.In addition, cell cultures from different mammals show variable susceptibilities to SARS-CoV-2, SARS-CoV, and MERS-CoV pseudotyped viruses. This implies that SARS-CoV-2 has the capacity to spillover to multiple species and establish natural reservoirs after minor or major adaptive evolutionary changes.Our results highlighted the potential for Thomas’s horseshoe bats, king horseshoe bats, green monkeys, and ferrets to serve as reservoir hosts for these coronaviruses. Specially, primary cells cultures derived from Thomas’s horseshoe bat and the king horseshoe bat were found to be more sensitive to VSVΔG*-SARS2 than human cell lines. This is important, as horseshoe bats (Rhinolophidae) are considered as a reservoir for many zoonotic viruses and they are assumed to be generally tolerant to infection.However, very rare sarbecoviruses have been reported in both Thomas’s horseshoe bats and the king horseshoe bats, but not in other horseshoe bats (e.g., Chinese horseshoe bats) [86].This not only raises the concern that SARS-CoV-2 might spillover from humans to these two bat species, but also prioritizes the surveillance of virus intolerance in horseshoe bats.(SNIP)Finally, our results highlight the fact that cell culture models represent an important method of understanding cross-species transmission of sarbecoviruses by identifying the spectrum of mammalian hosts that are susceptible to SARS-CoV-2, SARS-CoV, and MERS-CoV and their variants.
For more on the detection of SARS-CoV-2 in wildlife, you may wish to revisit these recent blogs:
The Lancet Microbe: Ecology of SARS-CoV-2 in the Post-Pandemic Era
Preprint: SARS-CoV-2 Exposure in Norwegian rats (Rattus norvegicus) from New York City
Nature: Divergent SARS-CoV-2 Variant Emerges in White-tailed Deer with Deer-to-Human Transmission (Revisited)
Preprint: Wildlife Exposure to SARS-CoV-2 Across a Human Use Gradient