# 5675
All that is required to spark a pandemic is for a novel influenza virus to emerge that mankind has little or no resistance to, for it to cause significant morbidity and mortality, and for it to adapt to human physiology so that it transmits efficiently.
The H5N1 virus fulfills these first two criteria, but fails on the third.
For now, anyway.
But there are many other influenza viruses circulating in birds, pigs, and other species that have the potential to either mutate - or more likely - reassort (swap gene segments with another flu strain) and adapt to human hosts.
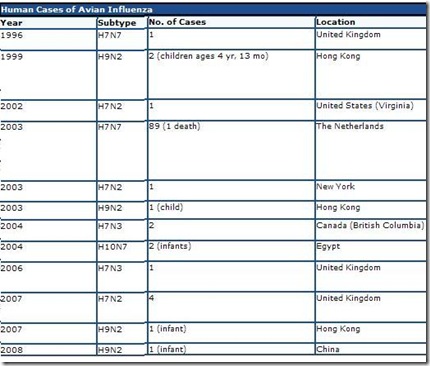
Below you’ll find a chart lifted and edited from CIDRAP’s excellent overview Avian Influenza (Bird Flu): Implications for Human Disease showing non-H5N1 avian flu infections in humans over the past decade.
Since surveillance is – at best - haphazard (or even non-existent) in many parts of the world, how often these types of novel infections really occur is unknown.
Despite rare known human infections, these viruses need to acquire genetic changes before they could spark a pandemic. Which is why we concern ourselves with their ability to reassort with other – already humanized – flu strains.
Reassortment (or Shift), happens when two different influenza viruses co-infect the same host and swap genetic material.

Influenza A viruses have 8 gene segments (PB2, PB1, PA, HA, NP, NA, M1, M2, NS1, NS2)
.
Which means that any two compatible influenza viruses could conceivably – and under the right conditions – generate 256 different combinations by swapping one or more of their 8 (potentially) interchangeable gene segments.
The key words being “under the right conditions”.
Last February in PNAS: Reassortment Of H1N1 And H9N2 Avian viruses we saw research from Chinese scientists that created – using reverse genetics – 128 reassorted viruses from the avian H9N2 virus and the (formerly pandemic) H1N1 virus.
In mouse testing, they found half of the hybrid viruses were biologically `fit’ as far as replication goes, and 8 hybrids were significantly more pathogenic than either of their parental viruses.
Today, again from PNAS, we have a new study that once again looks at the reassortment potential of the avian H9N2 virus and H1N1.
This time, research was done using ferrets, whose respiratory physiology is closer to human than are mice.
Compatibility of H9N2 avian influenza surface genes and 2009 pandemic H1N1 internal genes for transmission in the ferret model
J. Brian Kimble, Erin Sorrell, Hongxia Shao, Philip L. Martin, and Daniel Roberto Perez
Abstract
In 2009, a novel H1N1 influenza (pH1N1) virus caused the first influenza pandemic in 40 y. The virus was identified as a triple reassortant between avian, swine, and human influenza viruses, highlighting the importance of reassortment in the generation of viruses with pandemic potential.
Previously, we showed that a reassortant virus composed of wild-type avian H9N2 surface genes in a seasonal human H3N2 backbone could gain efficient respiratory droplet transmission in the ferret model.
Here we determine the ability of the H9N2 surface genes in the context of the internal genes of a pH1N1 virus to efficiently transmit via respiratory droplets in ferrets. We generated reassorted viruses carrying the HA gene alone or in combination with the NA gene of a prototypical H9N2 virus in the background of a pH1N1 virus.
Four reassortant viruses were generated, with three of them showing efficient respiratory droplet transmission. Differences in replication efficiency were observed for these viruses; however, the results clearly indicate that H9N2 avian influenza viruses and pH1N1 viruses, both of which have occasionally infected pigs, have the potential to reassort and generate novel viruses with respiratory transmission potential in mammals.
The entire study is available online, and open access.
As noted in the abstract above, these authors had previously successfully created laboratory reassortments between seasonal H3N2 and H9N2.
The fact that these hybrid viruses can be created in the laboratory doesn’t automatically mean they would be generated in the field by a co-infected host.
Only that it is possible.
And with 256 possible combinations, these 4 hybrids might not even represent the most `fit’ reassortments.
But research like this continues to show the potential for the H9N2 virus to move towards a more `humanized’ pathogen. And with H1N1 and H9N2 both known to be circulating in pigs in Asia, there are ample opportunities for them to co-infect the same host.
A few notable H9N2 stories from the past include:
- In December 2008 I ran a blog featuring an interview in which world famous Hong Kong virologist Malik Peiris cautioned that the H9N2 virus may be circulating far more commonly than we believe. Revisiting A Malik Peiris Interview On H9N2
- In January of 2010, in H9N2: The Other Bird Flu Threat, I wrote about the World Health Organization recommending the creation of a candidate vaccine virus for H9N2. According to the latest vaccine update from the WHO, work continues on that candidate vaccine virus.
- And just this past November, in Study: The Continuing Evolution Of Avian H9N2, we looked at computer modeling that suggested that the H9N2 virus moving towards becoming more `human-adapted’.
Unlike the H7 and H5 avian flu strains, poultry (and swine) infections by the H9N2 virus are not required to be reported to the OIE.
As we saw in 2009, sometimes a pandemic virus will emerge from an unexpected source, and with a surprising lineage. While the world was waiting for an H5 bird flu to emerge from Asia, we were blindsided by a H1N1 swine flu from North America.
All of which highlights the importance of establishing better global surveillance of humans, and farm animals, for the next emerging influenza virus.
Regardless of its strain.