Photo Credit – FAO
# 9514
We’ve a nifty piece of avian flu research which appears this week in PNAS, that looks at the continued evolution of H9N2 in Chinese poultry, and how that may have prompted the emergence of the H7N9 virus during the spring of 2013.
A joint effort of several Chinese science institutions and the St. Jude Children's Research Hospital, this study features Jinhua Liu, Ph.D., of the College of Veterinary Medicine at the China Agricultural University, and Dr. Robert Webster as co-corresponding authors.
Regular readers of this blog are aware that the avian H9N2 virus – which has been rife in Asian poultry for the past couple of decades – has been a major contributor to the creation of new avian viruses. Of the viruses we are currently watching with the most concern – H5N1, H7N9, H5N6, and H10N8 – all share several important features (see Study: Sequence & Phylogenetic Analysis Of Emerging H9N2 influenza Viruses In China):
- They all first appeared in Mainland China
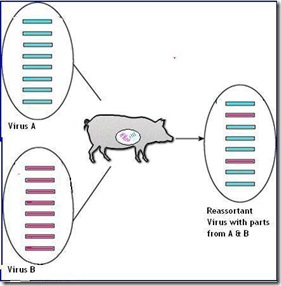
- They all have come about through viral reassortment in poultry
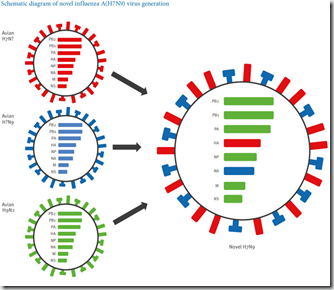
- And most telling of all, while their HA and NA genes differ - they all carry the internal genes from the avian H9N2 virus
Last January, The Lancet carried a report entitled Poultry carrying H9N2 act as incubators for novel human avian influenza viruses by Chinese researchers Di Liu a, Weifeng Shi b & George F Gao that warned:
Several subtypes of avian influenza viruses in poultry are capable of infecting human beings, and the next avian influenza virus that could cause mass infections is not known. Therefore, slaughter of poultry carrying H9N2—the incubators for wild-bird-origin influenza viruses—would be an effective strategy to prevent human beings from becoming infected with avian influenza.
Last May, in EID Journal: H7N9 As A Work In Progress, we looked at a study that found the H7N9 avian virus continues to reassort with local H9N2 viruses, making the H7N9 viruses that circulated in wave 2 genetically distinct from those that were seen during the 1st wave.
Although categorized by their two surface proteins (HA & NA) Influenza A viruses have 8 gene segments (PB2, PB1, PA, HA, NP, NA, M1, M2, NS1, NS2).
H7N9 carries 6 genes from H9N2 – Credit Eurosurveillance
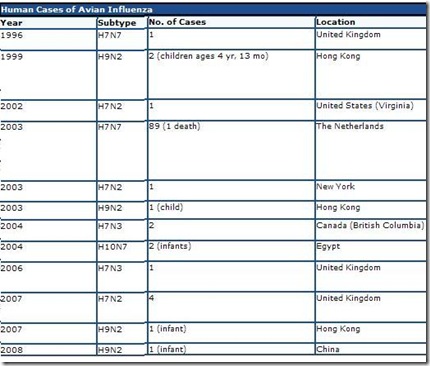
And while we have only seen a handful of human infections with H9N2 (see Hong Kong: Isolation & Treatment Of An H9N2 Patient), it is also true that in areas where this virus is most common, testing and surveillance for the virus is extremely limited. Like so many other novel viruses, we can only guess at is true burden in the human population.
While it’s impact on emerging avian flu viruses has been well documented, the reasons behind it’s influence have been less than clear.
Today, however, we have research that shows a new, better adapted genotype (G57) of the H9N2 virus has emerged – one that evades the poultry vaccines currently in use – and that it has become widespread among vaccinated Chinese poultry since 2010.
This spread has provided more opportunities for reassortment, and so we see more avian viruses emerging.
First the Abstract from the study, followed by some excerpts from a St. Jude Children’s Research Hospital press release, after which I’ll return with a bit more.
Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus
Juan Pua,b,1, Shuoguo Wangc,1, Yanbo Yind,1, Guozhong Zhanga, Robert A. Carterc, Jinliang Wanga, Guanlong Xua, Honglei Suna, Min Wanga, Chu Wena, Yandi Weia, Dongdong Wangd, Baoli Zhue, Gordon Lemmonc, Yuannian Jiaoc, Susu Duanb, Qian Wanga, Qian Dua, Meng Suna, Jinnan Baoa, Yipeng Suna, Jixun Zhaoa, Hui Zhangf, Gang Wuc, Jinhua Liua,2, and Robert G. Websterb,2
Significance
The emergence of human infection with a novel H7N9 avian influenza reassortant in China raises a pandemic concern. However, it is not fully understood how these H9N2 chicken viruses facilitated the genesis of the novel H7N9 viruses. Here we show that a “fittest” genotype (G57) emerged with changed antigenicity and improved adaptability in chickens. It became predominant in vaccinated farm chickens and caused widespread outbreaks before the H7N9 virus emergence, increasing reassortment between H9N2 and other subtype viruses and finally providing all of their internal genes to the novel H7N9 viruses. The prevalence and variation of H9N2 influenza virus in farmed poultry could provide an important early warning of the emergence of novel reassortants with pandemic potential.
Abstract
The emergence of human infection with a novel H7N9 influenza virus in China raises a pandemic concern. Chicken H9N2 viruses provided all six of the novel reassortant’s internal genes. However, it is not fully understood how the prevalence and evolution of these H9N2 chicken viruses facilitated the genesis of the novel H7N9 viruses. Here we show that over more than 10 y of cocirculation of multiple H9N2 genotypes, a genotype (G57) emerged that had changed antigenicity and improved adaptability in chickens. It became predominant in vaccinated farm chickens in China, caused widespread outbreaks in 2010–2013 before the H7N9 viruses emerged in humans, and finally provided all of their internal genes to the novel H7N9 viruses. The prevalence and variation of H9N2 influenza virus in farmed poultry could provide an important early warning of the emergence of novel reassortants with pandemic potential.
Tracing evolution of chicken flu virus yields insight into origins of deadly H7N9 strain
Memphis, Tennessee, December 29, 2014An international research team has shown how changes in a flu virus that has plagued Chinese poultry farms for decades helped create the novel avian H7N9 influenza A virus that has sickened more than 375 people since 2013. The research appears in the current online early edition of the scientific journal Proceedings of the National Academy of Sciences.
The results underscore the need for continued surveillance of flu viruses circulating on poultry farms and identified changes in the H9N2 virus that could serve as an early warning sign of emerging flu viruses with the potential to trigger a pandemic and global health emergency. The work focused on the H9N2 chicken virus, which causes egg production to drop and leaves chickens vulnerable to deadly co-infections. Scientists at St. Jude Children’s Research Hospital and the China Agricultural University, Beijing, led the study.
Researchers used whole genome sequencing to track the evolution of the H9N2 chicken virus between 1994 and 2013. The analysis involved thousands of viral sequences and showed that the genetic diversity of H9N2 viruses fell sharply in 2009. From 2010 through 2013 an H9N2 virus emerged as the predominant subtype thanks to its genetic makeup that allowed it to flourish despite widespread vaccination of chickens against H9N2 viruses.
Evidence in this study suggests the eruptions set the stage for the emergence of the H7N9 avian virus that has caused two outbreaks in humans since 2013, with 115 confirmed deaths. The H9N2 infected chickens likely served as the mixing vessel where H9N2 and other avian flu viruses from migratory birds and domestic ducks swapped genes, researchers noted. The resulting H7N9 virus included six genes from the H9N2.
"Sequencing the viral genome allowed us to track how H9N2 evolved across time and geography to contribute to the H7N9 virus that emerged as a threat to human health in 2013," said Robert Webster, Ph.D., a member of the St. Jude Department of Infectious Diseases. He and Jinhua Liu, Ph.D., of the College of Veterinary Medicine at the China Agricultural University, are co-corresponding authors.
"The insights gained from this collaboration suggest that tracking genetic diversity of H9N2 on poultry farms could provide an early warning of emerging viruses with the potential to spark a pandemic," Webster said.
If all of this sounds vaguely familiar, last April in Study: Sequence & Phylogenetic Analysis Of Emerging H9N2 influenza Viruses In China, we saw H9N2 viruses collected from two Chinese Provinces (Zhejiang & Guangdong) in late 2011 showed signs of evolving antigenically away from the vaccine strain, with a majority of isolates (14 out of 18) showing an amino acid change in the receptor binding site suggestive of an enhanced ability to bind to human receptor cells.
While poultry vaccination has been adopted by a number of countries to control their avian flu problems, there are distinct downsides to relying on vaccines for the control of avian viruses (for earlier blogs see OIE: Countries That Vaccinate Poultry Need An `Exit Strategy' & Food Insecurity, Economics, And The Control Of H7N9).
While vaccines can often protect poultry against illness - with increasingly diverse and rapidly evolving avian flu viruses - they can’t always prevent infection. Particularly if they aren’t updated often.
The end result being that subclinical infections can go undetected, viruses continue to circulate unnoticed, and new variants or reassortants continue to emerge. These failures are not limited to the H9N2 vaccines, as we’ve seen similar results with H5 vaccines as well (see EID Journal: Subclinical HPAI In Vaccinated Poultry – China & Egypt: A Paltry Poultry Vaccine).
There are, unfortunately, no simple solutions.
Countries like China, Indonesia, Vietnam, and Egypt – places where avian flu viruses are well entrenched in both commercial and home raised poultry - rely heavily on poultry vaccines. They view the immediate culling of infected birds, the standard control practice for most of the rest of the world, as being impractical and fear the social and economic impacts of such a policy.
The problem is, as new avian flu subtypes, clades, and sub clades emerge, the vaccines in use inevitably lose the battle, and over time that can lead to a snowball effect where the number of viruses in circulation escalate rapidly.
Given the recent emergence of H7N9, H10N8, H5N6, H5N8 and H5N3, one has to wonder just how much faster this snowball will roll over the next few years.