#17,120
Nine months ago, in Another Preprint On SARS-CoV-2 In White-Tailed Deer To Ponder, we looked at what was then the latest report on SARS-CoV-2 detected in North American White-Tailed Deer (WTD).
This one, however, had a bit of a twist; it described Highly divergent SARS-CoV-2 variants in WTD, and potential evidence of deer-to-human transmission.
Over the previous 8 months, we'd already seen:
Preprint: Evolutionary Trajectories of SARS-CoV-2 Alpha and Delta Variants in White-Tailed Deer in PennsylvaniaWhile the SARS-CoV-2 virus is remarkably well adapted to - and now endemic among - humans, it is also able to infect a number of other mammalian species (i.e. deer, mink, rodents, dogs, cats, etc.).
More Documented Spillovers Of COVID-19 Into North American Deer
Nature: SARS-CoV-2 Infection in Free-ranging White-tailed Deer
Two New Reports Find Widespread SARS-CoV-2 In North American Deer
This raises concerns that the virus could follow divergent evolutionary paths in non-human hosts, producing new, potentially dangerous variants that could eventually `spillback' into humans.
While controversial, there is also some evidence to suggest that the Omicron variant may have evolved after the virus jumped to mice or other rodents (see Evidence for a mouse origin of the SARS-CoV-2 Omicron variant), and then spilled back into humans.
Until earlier this year the viruses that have been detected in WTD had been quite similar to the variants currently (or previously) circulating in humans.
But in February 2022, a preprint on Evolutionary Trajectories of SARS-CoV-2 reported finding ". . .deer-derived alpha variants diverged significantly from those in humans, consistent with a distinctive evolutionary trajectory in deer."
That report was followed a week later by the above-mentioned preprint on the potential deer-to-human transmission. With thanks to my other reader (who sent me the link), we have the full, updated and peer-reviewed journal article, which was published this week in Nature.
This is a lengthy and detailed report, and I've only posted the abstract and a few excerpts. You'll want to follow the link and read it in its entirety. I'll have more after you return.
Bradley Pickering,Oliver Lung,Finlay Maguire,Peter Kruczkiewicz,Jonathon D. Kotwa,Tore Buchanan,Marianne Gagnier,Jennifer L. Guthrie,Claire M. Jardine,Alex Marchand-Austin,ArianeMassé,HeatherMcClinchey,Kuganya Nirmalarajah,Patryk Aftanas,Juliette Blais-Savoie,Hsien-Yao Chee, Emily Chien,Winfield Yim,Andra Banete,Bryan D. Griffin,Lily Yip,Melissa Goolia,Matthew Suderman,Mathieu Pinette,…Jeff Bowman
Nature Microbiology (2022)Cite this article
Abstract
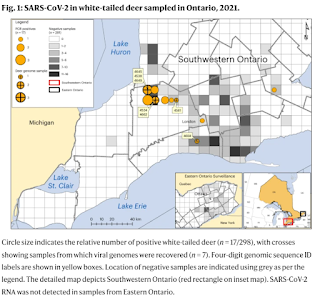
Wildlife reservoirs of broad-host-range viruses have the potential to enable evolution of viral variants that can emerge to infect humans. In North America, there is phylogenomic evidence of continual transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from humans to white-tailed deer (Odocoileus virginianus) through unknown means, but no evidence of transmission from deer to humans.
We carried out an observational surveillance study in Ontario, Canada during November and December 2021 (n = 300 deer) and identified a highly divergent lineage of SARS-CoV-2 in white-tailed deer (B.1.641). This lineage is one of the most divergent SARS-CoV-2 lineages identified so far, with 76 mutations (including 37 previously associated with non-human mammalian hosts). From a set of five complete and two partial deer-derived viral genomes we applied phylogenomic, recombination, selection and mutation spectrum analyses, which provided evidence for evolution and transmission in deer and a shared ancestry with mink-derived virus.
Our analysis also revealed an epidemiologically linked human infection. Taken together, our findings provide evidence for sustained evolution of SARS-CoV-2 in white-tailed deer and of deer-to-human transmission.
(SNIP)
It is unclear whether the initial spillover occurred directly from humans to deer, or an intermediate host such as mink or other yet undefined species was involved. The long branch length and period of unsampled evolution provide a number of possible scenarios (Fig. 6). The human-derived sample in the B.1.641 lineage lies within a relatively small number of deer-derived samples, rendering the precise relationship between the human- and deer-derived viruses uncertain (78% UFB). B.1.641 could represent a spillover into deer with a human spillback or the emergence of a virus reservoir in another wildlife species infecting both human and deer. However, the epidemiological data, evidence of infectious virus from deer, and the paucity of SARS-CoV-2 surveillance in white-tailed deer relative to human cases suggest spillover in deer followed by deer-to-human transmission is the most likely scenario (Fig. 6, scenario 1).
At this time, there is no evidence of recurrent deer-to-human or sustained human-to-human transmission of B.1.641. However, there has been considerable reduction in human and deer testing and genomic surveillance since the study samples were collected, and we cannot exclude the possibility of sustained transmission within or between these host populations. Enhanced surveillance is critical given human population density and mobility in the region, coupled with white-tailed deer population dynamics. In addition, rapid characterization of this new lineage from biological and epidemiological perspectives is critical to understanding viral transmission, immune evasion and disease in both wildlife and humans. Therefore, we assessed the ability of antibodies elicited following vaccination or infection to recognize and neutralize S and found that the mutations in B.1.641 do not have significant impact on S antigenicity (acknowledging the limited number of plasma tested) (Fig. 5). More work is needed to determine the potential roles of the mutations on spike functions, and to understand the pathogenesis and transmission phenotypes of this virus.
Secondary wildlife reservoirs have the potential to fundamentally alter the ecology of SARS-CoV-2. Our work underscores the need for a broad international One Health focus to identify new intermediate or reservoir hosts capable of driving sustained transmission and divergent viral evolution. An examination of human drivers of spillover and spillback and knock-on effects on wildlife and human health is urgently needed to identify, develop and implement mitigation strategies, beginning with reducing viral activity in humans.
Just as SARS-CoV-2 has evolved to become more transmissible in humans over the past 3 years, we've also seen evidence that some non-human species that were originally not believed susceptible to infection are now quite vulnerable.
Until about 18 months ago, rats and mice were thought largely immune to the virus, but in March of 2021 in PrePrint: The B1.351 and P.1 Variants Extend SARS-CoV-2 Host Range to Mice, we learned hat some SARS variants had overcome that barrier (also see Preprint: SARS-CoV-2 Infection in Domestic Rats After Transmission From Their Infected Owner).
Over the summer of 2021, in EID Journal: Peridomestic Mammal Susceptibility to SARS-CoV-2 Infection, we looked at the susceptibility of COVID in non-human species, either in farmed livestock or in the wild.
The authors wrote:
We show that several common peridomestic species, including deer mice, bushy-tailed woodrats, and striped skunks, are susceptible to infection and can shed the virus in respiratory secretions.In contrast, we demonstrate that cottontail rabbits, fox squirrels, Wyoming ground squirrels, black-tailed prairie dogs, house mice, and raccoons are not susceptible to SARS-CoV-2 infection.
While somewhat reassuring, the SARS-CoV-2 virus they challenged these animals with was from 2020 (WA1/2020WY96).
A far more recent report (see Preprint: Wildlife Exposure to SARS-CoV-2 Across a Human Use Gradient), found evidence of SARS-CoV-2 infection in wild species previously `cleared' of being carriers, including raccoons and cotton tailed rabbits.
The point being, the virus we are dealing with today is a far cry from the virus that emerged in 2019.
And with SARS-CoV-2 now established and circulating in multiple non-human species, the virus we may find ourselves dealing with a year or two from now could be even further removed from today's threat.
